The effect of molnupiravir and nirmatrelvir on SARS-CoV-2 genome diversity in severe models of COVID-19
- PMID: 40130852
- PMCID: PMC12053996
- DOI: 10.1128/spectrum.01829-24
The effect of molnupiravir and nirmatrelvir on SARS-CoV-2 genome diversity in severe models of COVID-19
Abstract
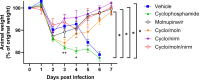
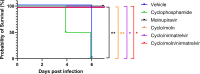
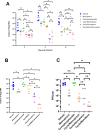
Immunocompromised individuals are susceptible to severe coronavirus disease 2019 and potentially contribute to the emergence of variants with altered pathogenicity due to persistent infection. This study investigated the impact of immunosuppression on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in K18-hACE2 mice and the effectiveness of antiviral treatments in this context during the first 7 days of infection. Mice were immunosuppressed using cyclophosphamide and infected with a B lineage of SARS-CoV-2. Molnupiravir and nirmatrelvir, alone and in combination, were administered, and viral load and viral sequence diversity were assessed. Treatment of infected but immunocompromised mice with both compounds either singly or in combination resulted in decreased viral loads and pathological changes compared to untreated animals. Treatment also abrogated infection of neuronal tissue. However, no consistent changes in the viral consensus sequence were observed, except for the emergence of the S:H655Y mutation. Molnupiravir, but not nirmatrelvir or immunosuppression alone, increased the transition/transversion ratio, representative of G > A and C > U mutations, and this increase was not altered by the co-administration of nirmatrelvir with molnupiravir. Notably, immunosuppression itself did not appear to promote the emergence of mutational characteristics of variants of concern (VOCs). Further investigations are warranted to fully understand the role of immunocompromised individuals in VOC development, especially by taking persistence into consideration, and to inform optimized public health strategies. It is more likely that immunodeficiency promotes viral persistence but does not necessarily lead to substantial consensus-level changes in the absence of antiviral selection pressure. Consistent with mechanisms of action, molnupiravir showed a stronger mutagenic effect than nirmatrelvir in this model.
Importance: The central aim of this study was to risk-assess the impact of administering a mutagenic antiviral compound, molnupiravir, to patients believed to already be at risk of generating increased viral diversity, which could have severe implications for antiviral resistance development. Combination therapy has a long history of mitigating antiviral resistance risk and was used in this study to demonstrate its potential usefulness in a severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) context. Animals treated with molnupiravir showed an increase in transition/transversion ratios over time, consistent with the drug's mechanism of action and a recent UK-wide phase II clinical trial assessing the efficacy of molnupiravir in humans. The addition of nirmatrelvir increased viral clearance, which in turn reduces the probability of viral persistence and rapid intra-host evolution of SARS-CoV-2.
Keywords: COVID-19; Paxlovid; SARS-CoV-2; immunocompromised; intra-host evolution; molnupiravir; nirmatrelvir.
Conflict of interest statement
A.O. is a director of Tandem Nano Ltd and co-inventor of patents relating to drug delivery. A.O. has been co-investigator on funding received by the University of Liverpool from ViiV Healthcare and Gilead Sciences in the past 3 years unrelated to COVID-19. A.O. has received personal fees from Gilead and Assembly Biosciences in the past 3 years, also unrelated to COVID-19. J.P.S. has received funding from ENA Respiratory Pty Ltd, Bicycle Tx Ltd, and Infex Therapeutics Ltd unrelated to this study. R.P.-R. is an employee at TopMD Precision Medicine Ltd. No other conflicts are declared by the other authors.
Figures



Update of
-
The effect of molnupiravir and nirmatrelvir on SARS-CoV-2 genome diversity in severe models of COVID-19.bioRxiv [Preprint]. 2024 Dec 20:2024.02.27.582110. doi: 10.1101/2024.02.27.582110. bioRxiv. 2024. Update in: Microbiol Spectr. 2025 May 6;13(5):e0182924. doi: 10.1128/spectrum.01829-24. PMID: 38464327 Free PMC article. Updated. Preprint.
Similar articles
-
The effect of molnupiravir and nirmatrelvir on SARS-CoV-2 genome diversity in severe models of COVID-19.bioRxiv [Preprint]. 2024 Dec 20:2024.02.27.582110. doi: 10.1101/2024.02.27.582110. bioRxiv. 2024. Update in: Microbiol Spectr. 2025 May 6;13(5):e0182924. doi: 10.1128/spectrum.01829-24. PMID: 38464327 Free PMC article. Updated. Preprint.
-
Effect of molnupiravir on SARS-CoV-2 evolution in immunocompromised patients: a retrospective observational study.Lancet Microbe. 2024 May;5(5):e452-e458. doi: 10.1016/S2666-5247(23)00393-2. Epub 2024 Mar 22. Lancet Microbe. 2024. PMID: 38527471
-
Efficacy of late-onset antiviral treatment in immunocompromised hosts with persistent SARS-CoV-2 infection.J Virol. 2024 Sep 17;98(9):e0090524. doi: 10.1128/jvi.00905-24. Epub 2024 Aug 29. J Virol. 2024. PMID: 39207133 Free PMC article.
-
A tale of two drugs: Molnupiravir and Paxlovid.Mutat Res Rev Mutat Res. 2025 Jan-Jun;795:108533. doi: 10.1016/j.mrrev.2025.108533. Epub 2025 Feb 6. Mutat Res Rev Mutat Res. 2025. PMID: 39920989 Review.
-
Molnupiravir: First Approval.Drugs. 2022 Mar;82(4):455-460. doi: 10.1007/s40265-022-01684-5. Drugs. 2022. PMID: 35184266 Free PMC article. Review.
References
-
- Moore SC, Penrice-Randal R, Alruwaili M, Randle N, Armstrong S, Hartley C, Haldenby S, Dong X, Alrezaihi A, Almsaud M, et al. . 2020. Amplicon-based detection and sequencing of SARS-CoV-2 in nasopharyngeal swabs from patients with COVID-19 and identification of deletions in the viral genome that encode proteins involved in interferon antagonism. Viruses 12:1164. doi:10.3390/v12101164 - DOI - PMC - PubMed
-
- Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, Sheffield COVID-19 Genomics Group, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC. 2020. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 Virus. Cell 182:812–827. doi:10.1016/j.cell.2020.06.043 - DOI - PMC - PubMed
-
- Goldswain H, Dong X, Penrice-Randal R, Alruwaili M, Shawli GT, Prince T, Williamson MK, Raghwani J, Randle N, Jones B, et al. . 2023. The P323L substitution in the SARS-CoV-2 polymerase (NSP12) confers a selective advantage during infection. Genome Biol 24. doi:10.1186/s13059-023-02881-5 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- R01 AI134091/AI/NIAID NIH HHS/United States
- IZSEZ0 213289/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung
- R24 AI118397/AI/NIAID NIH HHS/United States
- 75F40120C00085/U.S. Food and Drug Administration
- BB/R00904X/1, BB/R018863/1, BB/N022505/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R01AI134091, R24AI118397/NH/NIH HHS/United States
- 222489/Z/21/Z/WT_/Wellcome Trust/United Kingdom
- EP/R024804/1, EP/S012265/1/Wellcome EPSRC Centre for Medical Engineering
- WT_/Wellcome Trust/United Kingdom
- MR/W005611/1, MR/Y004205/1, MR/R010145/1, MR/W021641/1, PA6162_G2P2-2023/MRC_/Medical Research Council/United Kingdom
- TS/W022648/1/Innovate UK
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

