Genomic and structural insights into Jyvaskylavirus, the first giant virus isolated from Finland
- PMID: 40131223
- PMCID: PMC11936420
- DOI: 10.7554/eLife.103492
Genomic and structural insights into Jyvaskylavirus, the first giant virus isolated from Finland
Abstract
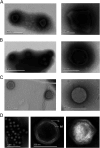
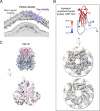
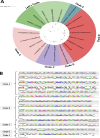
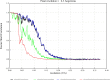
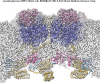
Giant viruses of protists are a diverse and likely ubiquitous group of organisms. Here, we describe Jyvaskylavirus, the first giant virus isolated from Finland. This clade B marseillevirus was found in Acanthamoeba castellanii from a composting soil sample in Jyväskylä, Central Finland. Its genome shares similarities with other marseilleviruses. Helium ion microscopy and electron microscopy of infected cells unraveled stages of the Jyvaskylavirus life cycle. We reconstructed the Jyvaskylavirus particle to 6.3 Å resolution using cryo-electron microscopy. The ~2500 Å diameter virion displays structural similarities to other Marseilleviridae giant viruses. The capsid comprises of 9240 copies of the major capsid protein, encoded by open reading frame (ORF) 184, which possesses a double jellyroll fold arranged in trimers forming pseudo-hexameric capsomers. Below the capsid shell, the internal membrane vesicle encloses the genome. Through cross-structural and -sequence comparisons with other Marseilleviridae using AI-based software in model building and prediction, we elucidated ORF142 as the penton protein, which plugs the 12 vertices of the capsid. Five additional ORFs were identified, with models predicted and fitted into densities that either cap the capsomers externally or stabilize them internally. The isolation of Jyvaskylavirus suggests that these viruses may be widespread in the boreal environment and provide structural insights extendable to other marseilleviruses.
Keywords: NCLDV; giant viruses; infectious disease; marseilleviruses; microbiology; molecular biophysics; structural biology; viruses.
Plain language summary
Viruses are everywhere. Some viruses can cause illness in humans, which has led to people often viewing them as threats. But most viruses are harmless to humans. Many viruses target microbes instead of humans. These viruses likely play essential roles in maintaining a healthy balance in ecosystems by preventing overpopulation of their target microbes. Yet, scientists know very little about most viruses and their role in ecosystems. Scientists have recently discovered a special group of giant viruses that target microscopic creatures called amoebas. The giant viruses that target them are much larger than most viruses and have some other unique features, such as having large genomes. A tough outer coating provides the structural reinforcement necessary to support their large size. Most of the giant viruses identified so far have been discovered in Europe and South America. A few have been found in North Africa, India, Japan and Siberia. But scientists still do not know how many giant viruses there are and where else they exist around the world. They are also still learning about the structures that allow these giant viruses to be so large. Almeida et al. describe the first giant virus ever discovered in Finland. In the experiments, the researchers mixed a type of amoeba called Acanthamoeba castellanii with environmental samples and monitored the amoebas for infection. The experiments revealed a giant amoeba-infecting virus in a compost sample. Almeida et al. named it Jyvaskylavirus after the city from which the compost sample came. The investigators then sequenced the virus’ DNA and used a cutting-edge imaging tool called cryogenic electron microscopy and artificial intelligence to determine details of the viral structure. This detailed structure of Jyvaskylavirus provided information about the structure of giant viruses and key structural proteins, which may also benefit scientists studying other kinds of viruses. The experiments confirm that giant viruses are a part of Finland’s boreal forest ecosystem, extending the known range of these unusual viruses. More studies are needed to identify the full range of giant viruses worldwide and to understand their role in ecosystems.
© 2024, Almeida et al.
Conflict of interest statement
GA, IA, Bd, ML, JA, JA, DZ, JR, NA, LS No competing interests declared
Figures






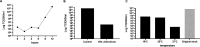




Update of
- doi: 10.1101/2024.10.16.618641
- doi: 10.7554/eLife.103492.1
- doi: 10.7554/eLife.103492.2
Similar articles
-
Complex Membrane Remodeling during Virion Assembly of the 30,000-Year-Old Mollivirus Sibericum.J Virol. 2019 Jun 14;93(13):e00388-19. doi: 10.1128/JVI.00388-19. Print 2019 Jul 1. J Virol. 2019. PMID: 30996095 Free PMC article.
-
Subnanometer structure of medusavirus capsid during maturation using cryo-electron microscopy.J Virol. 2024 Sep 17;98(9):e0043624. doi: 10.1128/jvi.00436-24. Epub 2024 Aug 28. J Virol. 2024. PMID: 39194243 Free PMC article.
-
Cryo-EM reconstruction of the Cafeteria roenbergensis virus capsid suggests novel assembly pathway for giant viruses.Sci Rep. 2017 Jul 14;7(1):5484. doi: 10.1038/s41598-017-05824-w. Sci Rep. 2017. PMID: 28710447 Free PMC article.
-
Structure and physiology of giant DNA viruses.Curr Opin Virol. 2021 Aug;49:58-67. doi: 10.1016/j.coviro.2021.04.012. Epub 2021 May 26. Curr Opin Virol. 2021. PMID: 34051592 Review.
-
Mimivirus: leading the way in the discovery of giant viruses of amoebae.Nat Rev Microbiol. 2017 Apr;15(4):243-254. doi: 10.1038/nrmicro.2016.197. Epub 2017 Feb 27. Nat Rev Microbiol. 2017. PMID: 28239153 Free PMC article. Review.
Cited by
-
Conserved marseilleviruses harboring diverse antibiotic resistance genes isolated from the Yangtze river Delta and the Pearl river delta, China.Sci Rep. 2025 Mar 27;15(1):10663. doi: 10.1038/s41598-025-94967-2. Sci Rep. 2025. PMID: 40148447 Free PMC article.
References
-
- Abrahão J, Silva L, Silva LS, Khalil JYB, Rodrigues R, Arantes T, Assis F, Boratto P, Andrade M, Kroon EG, Ribeiro B, Bergier I, Seligmann H, Ghigo E, Colson P, Levasseur A, Kroemer G, Raoult D, La Scola B. Tailed giant Tupanvirus possesses the most complete translational apparatus of the known virosphere. Nature Communications. 2018;9:749. doi: 10.1038/s41467-018-03168-1. - DOI - PMC - PubMed
-
- Abramson J, Adler J, Dunger J, Evans R, Green T, Pritzel A, Ronneberger O, Willmore L, Ballard AJ, Bambrick J, Bodenstein SW, Evans DA, Hung C-C, O’Neill M, Reiman D, Tunyasuvunakool K, Wu Z, Žemgulytė A, Arvaniti E, Beattie C, Bertolli O, Bridgland A, Cherepanov A, Congreve M, Cowen-Rivers AI, Cowie A, Figurnov M, Fuchs FB, Gladman H, Jain R, Khan YA, Low CMR, Perlin K, Potapenko A, Savy P, Singh S, Stecula A, Thillaisundaram A, Tong C, Yakneen S, Zhong ED, Zielinski M, Žídek A, Bapst V, Kohli P, Jaderberg M, Hassabis D, Jumper JM. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature. 2024;630:493–500. doi: 10.1038/s41586-024-07487-w. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

