A deficient CP24 allele defines variation for dynamic nonphotochemical quenching and photosystem II efficiency in maize
- PMID: 40132112
- PMCID: PMC12018801
- DOI: 10.1093/plcell/koaf063
A deficient CP24 allele defines variation for dynamic nonphotochemical quenching and photosystem II efficiency in maize
Erratum in
-
Correction to: A deficient CP24 allele defines variation for dynamic nonphotochemical quenching and photosystem II efficiency in maize.Plant Cell. 2025 Jun 4;37(6):koaf158. doi: 10.1093/plcell/koaf158. Plant Cell. 2025. PMID: 40554693 Free PMC article. No abstract available.
Abstract
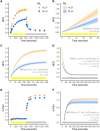
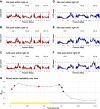
Maize (Zea mays L.) is a global crop species in which CO2 assimilation occurs via the C4 pathway. C4 photosynthesis is typically more efficient than C3 photosynthesis under warm and dry conditions; however, despite this inherent advantage, considerable variation remains in photosynthetic efficiency for C4 species that could be leveraged to benefit crop performance. Here, we investigate the genetic architecture of nonphotochemical quenching (NPQ) and photosystem II (PSII) efficiency using a combination of high-throughput phenotyping and quantitative trait loci (QTL) mapping in a field-grown Multi-parent Advanced Generation Inter-Cross (MAGIC) mapping population. QTL mapping was followed by the identification of putative candidate genes using a combination of genomics, transcriptomics, protein biochemistry, and targeted physiological phenotyping. We identified four genes with a putative causal role in the observed QTL effects. The highest confidence causal gene was found for a large effect QTL for photosynthetic efficiency on chromosome 10, which was underpinned by allelic variation in the expression of the minor PSII antenna protein light harvesting complex photosystem II subunit (LHCB6 or CP24), mainly driven by poor expression associated with the haplotype of the F7 founder line. The historical role of this line in breeding for early flowering time may suggest that the presence of this deficient allele could be enriched in temperate maize germplasm. These findings advance our understanding of the genetic basis of NPQ and PSII efficiency in C4 plants and highlight the potential for breeding strategies aimed at optimizing photosynthetic efficiency in maize.
© The Author(s) 2025. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures




References
-
- Andrews S. FastQC: a quality control tool for high throughput sequence data. v0.12.1 [2010]. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

