Nanoscale DNA tracing reveals the self-organization mechanism of mitotic chromosomes
- PMID: 40132578
- PMCID: PMC12127698
- DOI: 10.1016/j.cell.2025.02.028
Nanoscale DNA tracing reveals the self-organization mechanism of mitotic chromosomes
Abstract
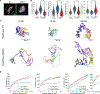
How genomic DNA is folded during cell division to form the characteristic rod-shaped mitotic chromosomes essential for faithful genome inheritance is a long-standing open question in biology. Here, we use nanoscale DNA tracing in single dividing cells to directly visualize how the 3D fold of genomic DNA changes during mitosis at scales from single loops to entire chromosomes. Our structural analysis reveals a characteristic genome scaling minimum of 6-8 megabases in mitosis. Combined with data-driven modeling and molecular perturbations, we can show that very large and strongly overlapping loops formed by condensins are the fundamental structuring principle of mitotic chromosomes. These loops compact chromosomes locally and globally to the limit set by chromatin self-repulsion. The characteristic length, density, and increasingly overlapping structure of mitotic loops we observe in 3D fully explain how the rod-shaped mitotic chromosome structure emerges by self-organization during cell division.
Keywords: cell division; chromatin tracing; chromosome compaction; condensins; genome organization; loop extrusion; mitosis.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

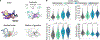
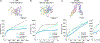


Update of
-
Nanoscale 3D DNA tracing reveals the mechanism of self-organization of mitotic chromosomes.bioRxiv [Preprint]. 2024 Oct 29:2024.10.28.620625. doi: 10.1101/2024.10.28.620625. bioRxiv. 2024. Update in: Cell. 2025 May 15;188(10):2656-2669.e17. doi: 10.1016/j.cell.2025.02.028. PMID: 39554202 Free PMC article. Updated. Preprint.
References
-
- Flemming W. (1882). Zellsubstanz, Kern und Zelltheilung (F.C.W. Vogel) 10.5962/bhl.title.168645. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

