Rapid and Integrated Bacterial Evolution Analysis unveils gene mutations and clinical risk of Klebsiella pneumoniae
- PMID: 40133255
- PMCID: PMC11937256
- DOI: 10.1038/s41467-025-58049-1
Rapid and Integrated Bacterial Evolution Analysis unveils gene mutations and clinical risk of Klebsiella pneumoniae
Abstract
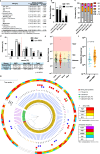
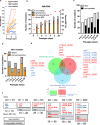
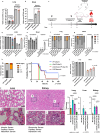
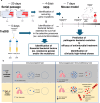
Bacteria continually evolve. Previous studies have evaluated bacterial evolution in retrospect, but this approach is based on only speculation. Cohort studies are reliable but require a long duration. Additionally, identifying which genetic mutations that have emerged during bacterial evolution possess functions of interest to researchers is an exceptionally challenging task. Here, we establish a Rapid and Integrated Bacterial Evolution Analysis (RIBEA) based on serial passaging experiments using hypermutable strains, whole-genome and transposon-directed sequencing, and in vivo evaluations to monitor bacterial evolution in a cohort for one month. RIBEA reveals bacterial factors contributing to serum and antimicrobial resistance by identifying gene mutations that occurred during evolution in the major respiratory pathogen Klebsiella pneumoniae. RIBEA also enables the evaluation of the risk for the progression and the development of invasive ability from the lung to blood and antimicrobial resistance. Our results demonstrate that RIBEA enables the observation of bacterial evolution and the prediction and identification of clinically relevant high-risk bacterial strains, clarifying the associated pathogenicity and the development of antimicrobial resistance at genetic mutation level.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests
Figures


References
MeSH terms
Substances
Grants and funding
- JP20ak0101118h0002, 22jk021004h0001, 23gm1610012h0001/Japan Agency for Medical Research and Development (AMED)
- 23gm1610012h0001, JP22wm0125008, JP23wm0125008, JP233fa627005/Japan Agency for Medical Research and Development (AMED)
- JP21H03622 and JP22K19416/MEXT | Japan Society for the Promotion of Science (JSPS)
LinkOut - more resources
Full Text Sources

