Leveraging large-scale biobank EHRs to enhance pharmacogenetics of cardiometabolic disease medications
- PMID: 40133288
- PMCID: PMC11937416
- DOI: 10.1038/s41467-025-58152-3
Leveraging large-scale biobank EHRs to enhance pharmacogenetics of cardiometabolic disease medications
Abstract
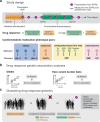
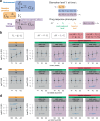
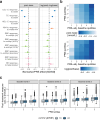
Electronic health records (EHRs) coupled with large-scale biobanks offer great promises to unravel the genetic underpinnings of treatment efficacy. However, medication-induced biomarker trajectories stemming from such records remain poorly studied. Here, we extract clinical and medication prescription data from EHRs and conduct GWAS and rare variant burden tests in the UK Biobank (discovery) and the All of Us program (replication) on ten cardiometabolic drug response outcomes including lipid response to statins, HbA1c response to metformin and blood pressure response to antihypertensives (N = 932-28,880). Our discovery analyses in participants of European ancestry recover previously reported pharmacogenetic signals at genome-wide significance level (APOE, LPA and SLCO1B1) and a novel rare variant association in GIMAP5 with HbA1c response to metformin. Importantly, these associations are treatment-specific and not associated with biomarker progression in medication-naive individuals. We also found polygenic risk scores to predict drug response, though they explained less than 2% of the variance. In summary, we present an EHR-based framework to study the genetics of drug response and systematically investigated the common and rare pharmacogenetic contribution to cardiometabolic drug response phenotypes in 41,732 UK Biobank and 14,277 All of Us participants.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: MCS has been consulting for 5 Prime Sciences at the time of the submission; however, this study was performed separately with no relationship to 5 Prime Sciences. The results and opinions expressed in this paper do not represent those of 5 Prime Sciences. The other authors declare that they have no competing interests.
Figures

Update of
-
Leveraging large-scale biobank EHRs to enhance pharmacogenetics of cardiometabolic disease medications.medRxiv [Preprint]. 2024 Apr 7:2024.04.06.24305415. doi: 10.1101/2024.04.06.24305415. medRxiv. 2024. Update in: Nat Commun. 2025 Mar 25;16(1):2913. doi: 10.1038/s41467-025-58152-3. PMID: 38633781 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- R35 GM153195/GM/NIGMS NIH HHS/United States
- U24 HG010615/HG/NHGRI NIH HHS/United States
- HG010615/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- 310030_189147/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (Swiss National Science Foundation)
LinkOut - more resources
Full Text Sources
Miscellaneous

