SCFA biotherapy delays diabetes in humanized gnotobiotic mice by remodeling mucosal homeostasis and metabolome
- PMID: 40133336
- PMCID: PMC11937418
- DOI: 10.1038/s41467-025-58319-y
SCFA biotherapy delays diabetes in humanized gnotobiotic mice by remodeling mucosal homeostasis and metabolome
Abstract
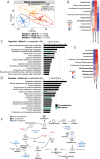
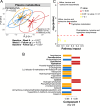
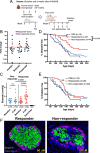
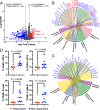
Type 1 diabetes (T1D) is linked to an altered gut microbiota characterized by reduced short-chain fatty acid (SCFA) production. Oral delivery of a SCFA-yielding biotherapy in adults with T1D was followed by increased SCFAs, altered gut microbiota and immunoregulation, as well as delaying diabetes in preclinical models. Here, we show that SCFA-biotherapy in humans is accompanied by remodeling of the gut proteome and mucosal immune homeostasis. Metabolomics showed arginine, glutamate, nucleotide and tryptophan metabolism were enriched following the SCFA-biotherapy, and found metabolites that correlated with glycemic control. Fecal microbiota transfer demonstrated that the microbiota of SCFA-responders delayed diabetes progression in humanized gnotobiotic mice. The protected mice increased similar metabolite pathways to the humans including producing aryl-hydrocarbon receptor ligands and reducing inflammatory mucosal immunity and increasing IgA production in the gut. These data demonstrate that a potent SCFA immunomodulator promotes multiple beneficial pathways and supports targeting the microbiota as an approach against T1D. Trial registration: Australia New Zealand Clinical Trials Registry ACTRN12618001391268.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: E.M. is an inventor on a patent WO2018027274A1 submitted by Monash University that covers methods and compositions for metabolites for the treatment and prevention of autoimmune disease related to this paper, and is the founder of ImmunoBiota Therapeutics Pty Ltd. All other authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

