Characterizing the tumor suppressor activity of FLCN in Birt-Hogg-Dubé syndrome cell models through transcriptomic and proteomic analysis
- PMID: 40133475
- PMCID: PMC12143978
- DOI: 10.1038/s41388-025-03325-z
Characterizing the tumor suppressor activity of FLCN in Birt-Hogg-Dubé syndrome cell models through transcriptomic and proteomic analysis
Abstract
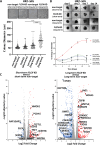
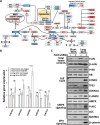
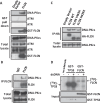
Birt-Hogg-Dubé syndrome (BHD) patients are uniquely susceptible to all renal tumor subtypes. However, the underlying mechanism of carcinogenesis is unclear. To study cancer development in BHD, we used human proximal kidney (HK2) cells and found that long-term folliculin (FLCN) knockdown was required to increase the tumorigenic potential of these cells, as evidenced by the formation of larger spheroids under nonadherent conditions. Transcriptomic and proteomic analyses revealed links between the FLCN, cell cycle control and DNA damage response (DDR) machinery. In addition, HK2 cells lacking FLCN had an altered transcriptome profile and enriched cell cycle control genes. G1/S cell cycle checkpoint signaling was compromised by increased protein levels of cyclin D1 (CCND1) and hyperphosphorylation of retinoblastoma 1 (RB1). A FLCN interactome screen revealed that FLCN binds to DNA-dependent protein kinase (DNA-PK). This novel interaction was reversed in an irradiation-responsive manner. Knockdown of FLCN in HK2 cells caused a marked increase in γH2AX and RB1 phosphorylation. The levels of both CCND1 and phosphorylated RB1 remained high during DNA damage, which was associated with defective cell cycle control caused by FLCN knockdown. Furthermore, Flcn-knockdown C. elegans were defective in cell cycle arrest caused by DNA damage. This work revealed that long-term FLCN loss and associated cell cycle defects in BHD patients could contribute to their increased risk of cancer.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing financial interests. While this study is funded in part by Health and Care Research Wales, the views expressed are those of the authors and not necessarily those of Health and Care Research Wales or the Welsh Government.
Figures



Update of
-
Characterizing the tumor suppressor activity of FLCN in Birt-Hogg-Dubé syndrome through transcriptiomic and proteomic analysis.Res Sq [Preprint]. 2024 Jun 26:rs.3.rs-4510670. doi: 10.21203/rs.3.rs-4510670/v1. Res Sq. 2024. Update in: Oncogene. 2025 Jun;44(23):1833-1843. doi: 10.1038/s41388-025-03325-z. PMID: 38978568 Free PMC article. Updated. Preprint.
References
-
- Birt AR, Hogg GR, Dubé WJ. Hereditary multiple fibrofolliculomas with trichodiscomas and acrochordons. Arch Dermatol. 1977;113:1674–7. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

