Characterizing resistant cellular states in nasopharyngeal carcinoma during EBV lytic induction
- PMID: 40133476
- PMCID: PMC12143974
- DOI: 10.1038/s41388-025-03341-z
Characterizing resistant cellular states in nasopharyngeal carcinoma during EBV lytic induction
Abstract
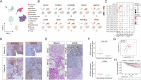
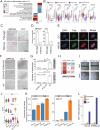
The pervasive occurrence of nasopharyngeal carcinoma (NPC) is intricately linked to Epstein-Barr virus (EBV) infection, making EBV and its associated pathways promising therapeutic targets for NPC and other EBV-related cancers. Lytic induction therapy, an emerging virus-targeted therapeutic strategy, capitalizes on the presence of EBV in tumor cells to specifically induce cytotoxicity against EBV-associated malignancies. Despite the expanding repertoire of compounds developed to induce EBV lytic reactivation, achieving universal induction across all infected cells remains elusive. The inherent heterogeneity of tumor cells likely contributes to this variability. In this study, we used the NPC43 cell line, an EBV-positive NPC in vitro model, and single-cell transcriptomics to characterize the diverse cellular responses to EBV lytic induction. Our longitudinal monitoring revealed a distinctive lytic induction non-responsive cellular state characterized by elevated expression of SOX2 and NTRK2. Cells in this state exhibit phenotypic similarities to cancer stem cells (CSCs), and we verified the roles of SOX2 and NTRK2 in manifesting these phenotypes. Our findings reveal a significant challenge for lytic induction therapy, as not all tumor cells are equally susceptible. These insights highlight the importance of combining lytic induction with therapies targeting CSC-like properties to enhance treatment efficacy for NPC and other EBV-associated cancers.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethical approval and consent to participate: All methods were performed in accordance with the “Chapter 9: Biological Safety” guidelines and regulations at HKUST. Written patient consents were obtained from all patients in this study according to institutional clinical research approval. The study protocol (2013.229) was approved by the Joint Chinese University of Hong Kong-New Territories East Cluster Clinical Research Ethics Committee at the Chinese University of Hong Kong, Hong Kong SAR.
Figures




References
-
- Wong KCW, Hui EP, Lo KW, Lam WKJ, Johnson D, Li L, et al. Nasopharyngeal carcinoma: an evolving paradigm. Nat Rev Clin Oncol. 2021;18:679–95. - PubMed
-
- Chua MLK, Wee JTS, Hui EP, Chan ATC. Nasopharyngeal carcinoma. Lancet. 2016;387:1012–24. - PubMed
-
- Chen YP, Chan ATC, Le QT, Blanchard P, Sun Y, Ma J. Nasopharyngeal carcinoma. Lancet. 2019;394:64–80. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

