Prospective, multicenter validation of a platform for rapid molecular profiling of central nervous system tumors
- PMID: 40133526
- PMCID: PMC12092301
- DOI: 10.1038/s41591-025-03562-5
Prospective, multicenter validation of a platform for rapid molecular profiling of central nervous system tumors
Abstract
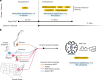
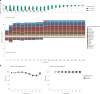
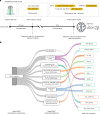
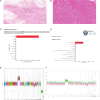
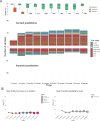
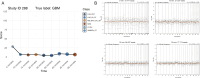
Molecular data integration plays a central role in central nervous system (CNS) tumor diagnostics but currently used assays pose limitations due to technical complexity, equipment and reagent costs, as well as lengthy turnaround times. We previously reported the development of Rapid-CNS2, an adaptive-sampling-based nanopore sequencing workflow. Here we comprehensively validated and further developed Rapid-CNS2 for intraoperative use. It now offers real-time methylation classification and DNA copy number information within a 30-min intraoperative window, followed by comprehensive molecular profiling within 24 h, covering the complete spectrum of diagnostically and therapeutically relevant information for the respective entity. We validated Rapid-CNS2 in a multicenter setting on 301 archival and prospective samples including 18 samples sequenced intraoperatively. To broaden the utility of methylation-based CNS tumor classification, we developed MNP-Flex, a platform-agnostic methylation classifier encompassing 184 classes. MNP-Flex achieved 99.6% accuracy for methylation families and 99.2% accuracy for methylation classes with clinically applicable thresholds across a global validation cohort of more than 78,000 frozen and formalin-fixed paraffin-embedded samples spanning five different technologies. Integration of these tools has the potential to advance CNS tumor diagnostics by providing broad access to rapid, actionable molecular insights crucial for personalized treatment strategies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: M. Sill, S.M.P., A.v.D., D.T.W.J., D.S. and F.S. are co-founders and shareholders of Heidelberg Epignostix GmbH. A.P. became a full-time employee of Heidelberg Epignostix GmbH in December 2024, while N.J. and M. Sill joined as full-time employees in July 2024, and D.S. became a part-time employee in November 2024. M.L. was a member of the Oxford Nanopore Technologies MinION access program and previously received free flow cells and sequencing reagents. M.L., S.H., H.L. and E.O.V.M. have received reimbursement for travel, accommodation and conference fees to speak at events organized by ONT. A.P., H.K., S.M.P., A.v.D., D.T.W.J., M.L., M. Sill and F.S. are inventors on a patent application related to a nanopore sequencing-based method for cancer characterization, filed by Deutsches Krebsforschungszentrum (DKFZ), Universität Heidelberg, and Oxford Nanopore Technologies PLC (patent application number: 18682016). The other authors declare no competing interests.
Figures




References
-
- Bouffet, E. et al. Dabrafenib plus trametinib in pediatric glioma with BRAF V600 mutations. N. Engl. J. Med.389, 1108–1120 (2023). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

