Haem biosynthesis regulates BCAA catabolism and thermogenesis in brown adipose tissue
- PMID: 40133548
- PMCID: PMC12116240
- DOI: 10.1038/s42255-025-01253-6
Haem biosynthesis regulates BCAA catabolism and thermogenesis in brown adipose tissue
Abstract
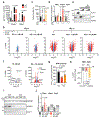
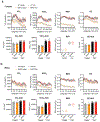
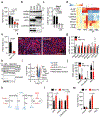
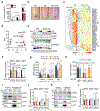
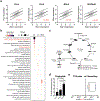
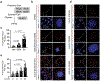
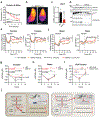
The distinctive colour of brown adipose tissue (BAT) is attributed to its high content of haem-rich mitochondria. However, the mechanisms by which BAT regulates intracellular haem levels remain largely unexplored. Here we demonstrate that haem biosynthesis is the primary source of haem in brown adipocytes. Inhibiting haem biosynthesis results in an accumulation of the branched-chain amino acids (BCAAs) valine and isoleucine, owing to a haem-associated metabolon that channels BCAA-derived carbons into haem biosynthesis. Haem synthesis-deficient brown adipocytes display reduced mitochondrial respiration and lower UCP1 levels than wild-type cells. Although exogenous haem supplementation can restore intracellular haem levels and mitochondrial function, UCP1 downregulation persists. This sustained UCP1 suppression is linked to epigenetic regulation induced by the accumulation of propionyl-CoA, a byproduct of disrupted haem synthesis. Finally, disruption of haem biosynthesis in BAT impairs thermogenic response and, in female but not male mice, hinders the cold-induced clearance of circulating BCAAs in a sex-hormone-dependent manner. These findings establish adipose haem biosynthesis as a key regulator of thermogenesis and sex-dependent BCAA homeostasis.
© 2025. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests: J.J.C. is a consultant for Thermo Fisher Scientific, 908 Devices and Seer. The other authors declare no competing interests.
Figures



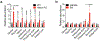

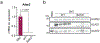


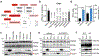

Update of
-
Heme biosynthesis regulates BCAA catabolism and thermogenesis in brown adipose tissue.bioRxiv [Preprint]. 2023 Nov 28:2023.11.28.568893. doi: 10.1101/2023.11.28.568893. bioRxiv. 2023. Update in: Nat Metab. 2025 May;7(5):1018-1033. doi: 10.1038/s42255-025-01253-6. PMID: 38076785 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- R35GM150899/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R35GM147014/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- PF-23-1070297-01-TBE/American Cancer Society (American Cancer Society, Inc.)
- TL1TR002375/U.S. Department of Health & Human Services | NIH | National Center for Advancing Translational Sciences (NCATS)
- TL1 TR002375/TR/NCATS NIH HHS/United States
- R35 GM147014/GM/NIGMS NIH HHS/United States
- T32 AG000213/AG/NIA NIH HHS/United States
- U54 DK110858/DK/NIDDK NIH HHS/United States
- UL1 TR002373/TR/NCATS NIH HHS/United States
- DK110858/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- P41GM108538/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R35 GM150899/GM/NIGMS NIH HHS/United States
- R01 CA286492/CA/NCI NIH HHS/United States
- 25PRE1374479/American Heart Association (American Heart Association, Inc.)
- P41 GM108538/GM/NIGMS NIH HHS/United States
- P30 DK020579/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

