Linking genomic and proteomic signatures to brain amyloid burden: insights from GR@ACE/DEGESCO
- PMID: 40133566
- PMCID: PMC11937198
- DOI: 10.1007/s10142-025-01581-6
Linking genomic and proteomic signatures to brain amyloid burden: insights from GR@ACE/DEGESCO
Abstract
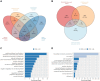
Alzheimer's disease (AD) is a complex disease with a strong genetic component, yet many genetic risk factors remain unknown. We combined genome-wide association studies (GWAS) on amyloid endophenotypes measured in cerebrospinal fluid (CSF) and positron emission tomography (PET) as surrogates of amyloid pathology, which may provide insights into the underlying biology of the disease. We performed a meta-GWAS of CSF Aβ42 and PET measures combining six independent cohorts (n = 2,076). Given the opposite beta direction of Aβ phenotypes in CSF and PET measures, only genetic signals showing opposite directions were considered for analysis (n = 376,599). We explored the amyloidosis signature in the CSF proteome using SOMAscan proteomics (ACE cohort, n = 1,008), connected it with GWAS loci modulating amyloidosis and performed an enrichment analysis of overlapping hits. Finally, we compared our results with a large meta-analysis using publicly available datasets in CSF (n = 13,409) and PET (n = 13,116). After filtering the meta-GWAS, we observed genome-wide significance in the rs429358-APOE locus and annotated nine suggestive hits. We replicated the APOE loci using the large CSF-PET meta-GWAS, identifying multiple AD-associated genes including the novel GADL1 locus. Additionally, we found 1,387 FDR-significant SOMAscan proteins associated with CSF Aβ42 levels. The overlap among GWAS loci and proteins associated with amyloid burden was minimal (n = 35). The enrichment analysis revealed mechanisms connecting amyloidosis with the plasma membrane's anchored component, synapse physiology and mental disorders that were replicated in the large CSF-PET meta-analysis. Combining CSF and PET amyloid GWAS with CSF proteome analyses may effectively elucidate causative molecular mechanisms behind amyloid mobilization and AD physiopathology.
Keywords: Aβ42; CSF biomarkers; GWAS; PET tomography; Proteome.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: In accordance with Spanish regulations for the biomedical research field, all the protocols of this study were approved by the Clinical Research Ethics Commission of the Hospital Clinic (Barcelona, Spain) for ACE cohort and the Clinical Research Ethics Commission of Cantabria (Spain) for Valdecilla cohort. This research followed the Declaration of Helsinki. All participants were informed about the procedures and objectives of this study by a neurologist before signing an informed consent. Moreover, data confidentiality and privacy of patients were protected as specified in applicable laws. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




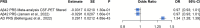

References
-
- Alcolea D, Pegueroles J, Muñoz L, Camacho V, López-Mora D, Fernández-León A, Le Bastard N, Huyck E, Nadal A, Olmedo V, Sampedro F, Montal V, Vilaplana E, Clarimón J, Blesa R, Fortea J, Lleó A (2019) Agreement of amyloid PET and CSF biomarkers for Alzheimer’s disease on Lumipulse. Ann Clin Trans Neurol 6(9):1815–1824. 10.1002/acn3.50873 - PMC - PubMed
-
- Alegret M, Valero S, Ortega G, Espinosa A, Sanabria A, Hernández I, Rodríguez O, Rosende-Roca M, Mauleón A, Vargas L, Martín E, Ruíz A, Tárraga L, Amariglio RE, Rentz DM, Boada M (2015) Validation of the Spanish version of the face name associative memory exam (S-FNAME) in cognitively normal older individuals. Arch Clin Neuropsychol 30(7):712–720. 10.1093/ARCLIN/ACV050 - PMC - PubMed
-
- Ali M, Archer DB, Gorijala P, Western D, Timsina J, Fernández MV, Wang TC, Satizabal CL, Yang Q, Beiser AS, Wang R, Chen G, Gordon B, Benzinger TLS, Xiong C, Morris JC, Bateman RJ, Karch CM, McDade E, … Cruchaga C (2023) Large multi-ethnic genetic analyses of amyloid imaging identify new genes for Alzheimer disease. Acta Neuropathol Commun 11(1):1–20. 10.1186/S40478-023-01563-4/FIGURES/4 - PMC - PubMed
-
- Alzheimer’s Association (2022) 2022 Alzheimer’s disease facts and figures: more than normal aging: understanding mild cognitive impairment. Alzheimers Dement 18(4):700–789 - PubMed
-
- Asada-Utsugi M, Uemura K, Noda Y, Kuzuya A, Maesako M, Ando K, Kubota M, Watanabe K, Takahashi M, Kihara T, Shimohama S, Takahashi R, Berezovska O, Kinoshita A (2011) N-cadherin enhances APP dimerization at the extracellular domain and modulates Aβ production. J Neurochem 119(2):354–363. 10.1111/J.1471-4159.2011.07364.X - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

