Metronidazole response profiles of Gardnerella species are congruent with phylogenetic and comparative genomic analyses
- PMID: 40133961
- PMCID: PMC11934483
- DOI: 10.1186/s13073-025-01446-4
Metronidazole response profiles of Gardnerella species are congruent with phylogenetic and comparative genomic analyses
Abstract
Background: Bacterial vaginosis (BV) affects 20-50% of reproductive-age female patients annually, arising when opportunistic pathogens outcompete healthy vaginal flora. Many patients fail to resolve symptoms with a course of metronidazole, the current first-line treatment for BV. Our study was designed to identify genomic variation associated with metronidazole resistance among strains of Gardnerella vaginalis spp. (GV), a genus of biogenic-amine-producing bacteria closely associated with BV pathogenesis, for the development of a companion molecular diagnostic.
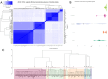
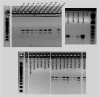
Methods: Whole-genome sequencing and comparative genomic metrics, including average nucleotide identity and GC content, were performed on a diverse set of 129 GV genomes to generate data for detailed taxonomic analyses. Pangenomic analyses were employed to construct a phylogenetic tree and cluster highly related strains within genospecies. G. vaginalis spp. clinical isolates within our collection were subjected to plate-based minimum inhibitory concentration (MIC) testing of metronidazole (n = 60) and clindamycin (n = 63). DECIPHER and MAFFT were used to identify genospecies-specific primers associated with antibiotic-resistance phenotypes. PCR-based analyses with these primers were used to confirm their specificity for the relevant genospecies.
Results: Eleven distinct genospecies based on standard ANI criteria were identified among the GV strains in our collection. Metronidazole MIC testing revealed six genospecies within a closely related phylogenetic clade contained only highly metronidazole-resistant strains (MIC ≥ 32 µg/mL) and suggested at least two mechanisms of metronidazole resistance within the eleven GV genospecies. All strains within the six highly metronidazole-resistant genospecies displayed susceptibility to clinically relevant clindamycin concentrations (MIC ≤ 2 µg/mL). A PCR-based molecular diagnostic assay was developed to distinguish between members of the metronidazole-resistant and mixed-response genospecies, which should be useful for determining the clade membership of various GV strains and could assist in the selection of appropriate antibiotic therapies for BV cases.
Conclusions: This study provides comparative genomic and phylogenetic evidence for eleven distinct genospecies within the genus Gardnerella vaginalis spp., and identifies genospecies-specific responses to metronidazole, the first-line treatment for BV. A companion molecular diagnostic assay was developed that is capable of identifying essentially all highly metronidazole-resistant strains that phylogenetically cluster together within the GV genospecies, which is informative for antibiotic treatment options.
Keywords: Antibiotic resistance; Bacterial vaginosis; Comparative genomics; Distributed genome hypothesis; Gardnerella vaginalis; Metronidazole resistance; Molecular diagnostic; Pangenome; Phylogenetics; Supragenome.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The Gardnerella vaginalis spp. isolates included in this study were made available from the biorepository of one of the authors of this study (SLH) at the University of Pittsburgh and Magee-Women’s Research Institute. The bacterial isolates were obtained from individuals enrolled in nine different clinical trials or observational cohort studies. All participants provided written informed consent prior to the collection of their samples and provided future use consent for use of their samples or sample remnants. The use of bacterial isolates in the present study does not constitute human subjects research because neither the repository holder nor the recipient of the isolates can link the isolates to any identifiable living individual. All links between the study participants, the bacterial isolates derived from their samples and their personal identifiers have been destroyed. Consent for publication: Not applicable. Competing interests: Authors KAI, JPE, and GDE have filed for a patent on the molecular diagnostic/primer sequences noted in the study. The remaining authors declare that they have no competing interests.
Figures




References
-
- Hillier SL. Diagnostic microbiology of bacterial vaginosis. Am J Obstet Gynecol. 1993;169:455–9. 10.1016/0002-9378(93)90340-o. - PubMed
-
- Sweet RL. Role of bacterial vaginosis in pelvic inflammatory disease. Clin Infect Dis. 1995;20(Suppl 2):S271–5. 10.1093/clinids/20.supplement_2.s271. - PubMed
-
- Soper DE. Bacterial vaginosis and postoperative infections. Am J Obstet Gynecol. 1993;169:467–9. 10.1016/0002-9378(93)90343-h. - PubMed
-
- Gravett MG, Hummel D, Eschenbach DA, Holmes KK. Preterm labor associated with subclinical amniotic fluid infection and with bacterial vaginosis. Obstet Gynecol. 1986;67:229–37. 10.1097/00006250-198602000-00013. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

