Subtelomeric repeat expansion in Hydractinia symbiolongicarpus chromosomes
- PMID: 40134021
- PMCID: PMC11934779
- DOI: 10.1186/s13100-025-00355-y
Subtelomeric repeat expansion in Hydractinia symbiolongicarpus chromosomes
Abstract
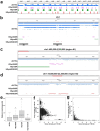
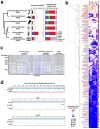
Despite the striking conservation of animal chromosomes, their repetitive element complements are vastly diverse. Only recently, high quality chromosome-level genome assemblies enabled identification of repeat compositions along a broad range of animal chromosomes. Here, utilizing the chromosome-level genome assembly of Hydractinia symbiolongicarpus, a colonial hydrozoan cnidarian, we describe an accumulation of a single 372 bp repeat unit in the subtelomeric regions. Based on the sequence divergence, its partial affinity with the Helitron group can be detected. This sequence is associated with a repeated minisatellite unit of about 150 bp. Together, they account for 26.1% of the genome (126 Mb of the 483 Mb). This could explain the genome size increase observed in H. symbiolongicarpus compared with other cnidarians, yet distinguishes this expansion from other large cnidarian genomes, such as Hydra vulgaris, where such localized propagation is absent. Additionally, we identify a derivative of an IS3EU-like DNA element accumulated at the putative centromeric regions. Our analysis further reveals that Helitrons generally comprise a large proportion of H. symbiolongicarpus (11.8%). We investigated Helitron presence and distributions across several cnidarian genomes. We find that in Nematostella vectensis, an anthozoan cnidarian, Helitron-like sequences were similarly accumulated at the subtelomeric regions. All these findings suggest that Helitron derivatives are prone to forming chromosomal extensions in cnidarians through local amplification in subtelomeric regions, driving variable genome expansions within the clade.
Keywords: Helitron; Hydractinia symbiolongicarpus; Cnidarian; Genome expansion; IS3EU; Rolling-circle transposon; Subtelomere; Tandem repeats; Transposable elements.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


References
-
- Simakov O, Bredeson J, Berkoff K, Marletaz F, Mitros T, Schultz DT et al. Deeply conserved synteny and the evolution of metazoan chromosomes. Sci Adv [Internet]. 2022;8:eabi5884. Available from: http://www.ncbi.nlm.nih.gov/pubmed/35108053 - PMC - PubMed
-
- Schultz DT, Haddock SHD, Bredeson JV, Green RE, Simakov O, Rokhsar DS. Ancient gene linkages support ctenophores as sister to other animals. Nature [Internet]. 2023;618:110–7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/37198475 - PMC - PubMed
-
- Schartl M, Woltering JM, Irisarri I, Du K, Kneitz S, Pippel M et al. The genomes of all lungfish inform on genome expansion and tetrapod evolution. Nature [Internet]. 2024;634:96–103. Available from: https://www.nature.com/articles/s41586-024-07830-1 - PMC - PubMed
-
- Meyer A, Schloissnig S, Franchini P, Du K, Woltering JM, Irisarri I et al. Giant lungfish genome elucidates the conquest of land by vertebrates. Nature [Internet]. 2021;590:284–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/33461212 - PMC - PubMed
-
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A et al. Sea Anemone Genome Reveals Ancestral Eumetazoan Gene Repertoire and Genomic Organization. Science (80-) [Internet]. 2007;317:86–94. Available from: https://www.science.org/doi/10.1126/science.1139158 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

