Correlative 3D imaging method for analysing lesion architecture in susceptible mice infected with Mycobacterium tuberculosis
- PMID: 40134379
- PMCID: PMC11972079
- DOI: 10.1242/dmm.052185
Correlative 3D imaging method for analysing lesion architecture in susceptible mice infected with Mycobacterium tuberculosis
Abstract
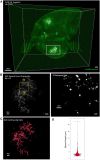
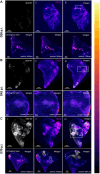
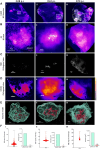
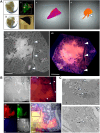
Tuberculosis (TB) is characterized by the formation of heterogeneous, immune-rich granulomas in the lungs. Host and pathogen factors contribute to this heterogeneity, but the molecular and cellular drivers of granuloma diversity remain inadequately understood owing to limitations in experimental techniques. In this study, we developed an approach that combines passive CLARITY (PACT)-based clearing with light-sheet fluorescence microscopy to visualize lesion architecture and lung involvement in Mycobacterium tuberculosis-infected C3HeB/FeJ mice. Three-dimensional rendering of post-mortem lungs revealed critical architectural features in lesion development that traditional thin-section imaging could not detect. Wild-type M. tuberculosis infection resulted in organized granulomas, with median sizes increasing to 3.74×108 µm3 and occupying ∼10% of the total lung volume by day 70 post-infection. In contrast, infection with the avirulent ESX-1 deletion mutant strain resulted in diffuse and sparsely organized CD11b recruitment (median size of 8.22×107 µm3), primarily located in the lung periphery and minimally involving the airways (0.23% of the total lung space). Additionally, we present a method for volumetric correlative light and electron microscopy, enabling tracking of individual immune cell populations within granulomas.
Keywords: CLARITY; Granuloma; Light-sheet fluorescence microscopy; Serial block face electron microscopy; Three-dimensional; Tuberculosis; Virulence.
© 2025. Published by The Company of Biologists.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures




References
-
- Clemmensen, H. S., Knudsen, N. P. H., Rasmussen, E. M., Winkler, J., Rosenkrands, I., Ahmad, A., Lillebaek, T., Sherman, D. R., Andersen, P. L. and Aagaard, C. (2017). An attenuated Mycobacterium tuberculosis clinical strain with a defect in ESX-1 secretion induces minimal host immune responses and pathology. Sci. Rep. 7, 46666. 10.1038/srep46666 - DOI - PMC - PubMed
-
- Cohen, S. B., Gern, B. H., Delahaye, J. L., Adams, K. N., Plumlee, C. R., Winkler, J. K., Sherman, D. R., Gerner, M. Y. and Urdahl, K. B. (2018). Alveolar macrophages provide an early mycobacterium tuberculosis niche and initiate dissemination. Cell Host Microbe 24, 439-446. 10.1016/j.chom.2018.08.001 - DOI - PMC - PubMed
-
- Cronan, M. R., Rosenberg, A. F., Oehlers, S. H., Saelens, J. W., Sisk, D. M., Smith, K. L. J., Lee, S. and Tobin, D. M. (2015). CLARITY and PACT-based imaging of adult zebrafish and mouse for whole-animal analysis of infections. Dis. Model. Mech. 8, 1643-1650. 10.1242/dmm.021394 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

