Decoding plasma cell maturation dynamics with BCMA
- PMID: 40134424
- PMCID: PMC11932843
- DOI: 10.3389/fimmu.2025.1539773
Decoding plasma cell maturation dynamics with BCMA
Abstract
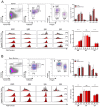
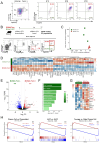
Plasma cells provide protective antibodies following an infection or vaccination. A network of intrinsic and extrinsic factors fine-tunes the generation of a heterogenous plasma cell pool with varying metabolic requirements, transcriptional profiles and lifespans. Among these, the B cell maturation antigen (BCMA) has been implicated in the APRIL-mediated survival of long-lived plasma cells. To characterize the terminal maturation of plasma cells, we constructed a BCMA reporter mouse (BCMA:Tom) that exclusively labeled antibody-secreting cells and revealed that BCMA:Tom expression varied by IgH isotype and increased with plasma cell maturity. The BCMA reporter, used alongside the Blimp1-GFP reporter, also allowed detailed tracking of plasma cell development and highlighted the importance of the in vivo microenvironment to complete plasma cell maturation. Therefore, the BCMA:Tom reporter mouse provides a valuable tool for tracking plasma cell development and maturation with flow cytometry or advanced imaging techniques, enabling a deeper understanding of the mechanisms regulating plasma cell heterogeneity and longevity.
Keywords: BCMA (TNFRSF17); antibody-secreting cells (ASC); bone marrow; plasma cells; spleen; survival; thymus.
Copyright © 2025 Schulz, Menzel, Wittner, Ulbricht, Grofe, Roth, Mann-Nüttel, Scheu, Kueh, Jäck, Herold, Hauser, Pracht, Schuh and Jäck.
Conflict of interest statement
The authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

