GNLY+CD8+ T cells bridge premature aging and persistent inflammation in people living with HIV
- PMID: 40135938
- PMCID: PMC11948365
- DOI: 10.1080/22221751.2025.2466695
GNLY+CD8+ T cells bridge premature aging and persistent inflammation in people living with HIV
Abstract
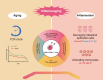
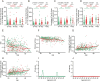
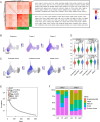
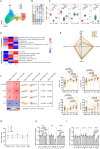
People living with HIV (PLWH) exhibit accelerated aging, characterized by systemic inflammation, termed "inflammaging." While T-cell expansion is prevalent in PLWH, its connection to inflammaging remains unclear. In this study, we analyzed the TCRβ repertoire of 257 healthy controls (HC) and 228 PLWH, revealing pronounced T cell clonal expansion in PLWH. The expansion was only partially reversed following antiretroviral therapy (ART) and closely associated with ART duration, CD4+ T and CD8+ T cell counts and the CD4/CD8 ratio. TCR-based age modeling showed a continuous accelerated trajectory of aging in PLWH, especially in younger individuals, in stark contrast to the nonlinear aging acceleration pattern seen in HC. Furthermore, using single-cell RNA combined TCR sequencing and in vitro experiments, we identified GNLY+CD8+ T cells as the primary population driving clonal expansion and maintenance in PLWH. These cells are characterized by high cytotoxicity and low exhaustion and are activated by interleukin-15 (IL-15) in vitro. Notably, GNLY+CD8+ T cells predominantly express the pro-inflammatory 15 kDa form of granulysin(GNLY). The supernatant from IL-15-stimulated CD8+ T cells induces monocytes to secrete inflammatory factors and disrupts the integrity of intestinal epithelial cells, which can be partially restored by the anti-GNLY antibodies. These findings identify GNLY+CD8+ T cells as the central drivers of persistent clonal expansion, highlighting their crucial role for mitigating inflammaging in PLWH.
Keywords: GNLY+CD8+ T cells; HIV; TCR repertoire; clonal expansion; inflammaging.
Conflict of interest statement
Dr. Zhixi Zhang is the founder of Chengdu ExAb Biotechnology Ltd. All financial interests are unrelated to this study. The remaining authors declare no competing interests.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
