Neutralization and spike stability of JN.1-derived LB.1, KP.2.3, KP.3, and KP.3.1.1 subvariants
- PMID: 40136024
- PMCID: PMC12077133
- DOI: 10.1128/mbio.00464-25
Neutralization and spike stability of JN.1-derived LB.1, KP.2.3, KP.3, and KP.3.1.1 subvariants
Abstract
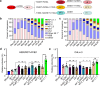
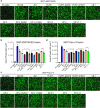
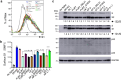
During the summer of 2024, coronavirus disease 2019 (COVID-19) cases surged globally, driven by variants derived from JN.1 subvariants of severe acute respiratory syndrome coronavirus 2 that feature new mutations, particularly in the N-terminal domain (NTD) of the spike protein. In this study, we report on the neutralizing antibody (nAb) escape, infectivity, fusion, and spike stability of these subvariants-LB.1, KP.2.3, KP.3, and KP.3.1.1. Our findings demonstrate that all of these subvariants are highly evasive of nAbs elicited by the bivalent mRNA vaccine, the XBB.1.5 monovalent mumps virus-based vaccine, or from infections during the BA.2.86/JN.1 wave. This reduction in nAb titers is primarily driven by a single serine deletion (DelS31) in the NTD of the spike, leading to a distinct antigenic profile compared to the parental JN.1 and other variants. We also found that the DelS31 mutation decreases pseudovirus infectivity in CaLu-3 cells, which correlates with impaired cell-cell fusion. Additionally, the spike protein of DelS31 variants appears more conformationally stable, as indicated by reduced S1 shedding both with and without stimulation by soluble ACE2 and increased resistance to elevated temperatures. Molecular modeling suggests that DelS31 enhances the NTD-receptor-binding domain (RBD) interaction, favoring the RBD down conformation and reducing accessibility to ACE2 and specific nAbs. Moreover, DelS31 introduces an N-linked glycan at N30, shielding the NTD from antibody recognition. These findings underscore the role of NTD mutations in immune evasion, spike stability, and viral infectivity, highlighting the need to consider DelS31-containing antigens in updated COVID-19 vaccines.IMPORTANCEThe emergence of novel severe acute respiratory syndrome coronavirus 2 variants continues to pose challenges for global public health, particularly in the context of immune evasion and viral stability. This study identifies a key N-terminal domain (NTD) mutation, DelS31, in JN.1-derived subvariants that enhances neutralizing antibody escape while reducing infectivity and cell-cell fusion. The DelS31 mutation stabilizes the spike protein conformation, limits S1 shedding, and increases thermal resistance, which possibly contribute to prolonged viral persistence. Structural analyses reveal that DelS31 enhances NTD-receptor-binding domain interactions by introducing glycan shielding, thus decreasing antibody and ACE2 accessibility. These findings emphasize the critical role of NTD mutations in shaping viral evolution and immune evasion, underscoring the urgent need for updated coronavirus disease 2019 vaccines that account for these adaptive changes.
Keywords: COVID-19 vaccine; DelS31 mutation; JN.1 subvariants; SARS-CoV-2; cell-cell fusion; immune evasion; spike protein stability.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Update of
-
Neutralization and Stability of JN.1-derived LB.1, KP.2.3, KP.3 and KP.3.1.1 Subvariants.bioRxiv [Preprint]. 2024 Sep 5:2024.09.04.611219. doi: 10.1101/2024.09.04.611219. bioRxiv. 2024. Update in: mBio. 2025 May 14;16(5):e0046425. doi: 10.1128/mbio.00464-25. PMID: 39282390 Free PMC article. Updated. Preprint.
References
-
- CDC . 2024. COVID data tracker, variant proportions. Centers for Disease Control
-
- Gangavarapu K, Latif AA, Mullen JL, Alkuzweny M, Hufbauer E, Tsueng G, Haag E, Zeller M, Aceves CM, Zaiets K, Cano M, Zhou X, Qian Z, Sattler R, Matteson NL, Levy JI, Lee RTC, Freitas L, Maurer-Stroh S, GISAID Core and Curation Team, Suchard MA, Wu C, Su AI, Andersen KG, Hughes LD. 2023. Outbreak.info genomic reports: scalable and dynamic surveillance of SARS-CoV-2 variants and mutations. Nat Methods 20:512–522. doi:10.1038/s41592-023-01769-3 - DOI - PMC - PubMed
-
- Planas D, Staropoli I, Michel V, Lemoine F, Donati F, Prot M, Porrot F, Guivel-Benhassine F, Jeyarajah B, Brisebarre A, Dehan O, Avon L, Bolland WH, Hubert M, Buchrieser J, Vanhoucke T, Rosenbaum P, Veyer D, Péré H, Lina B, Trouillet-Assant S, Hocqueloux L, Prazuck T, Simon-Loriere E, Schwartz O. 2024. Distinct evolution of SARS-CoV-2 Omicron XBB and BA.2.86/JN.1 lineages combining increased fitness and antibody evasion. Nat Commun 15:2254. doi:10.1038/s41467-024-46490-7 - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
- P23OA000000C019/U.S. Department of Agriculture (USDA)
- U54 CA260582/CA/NCI NIH HHS/United States
- U01 AI173348/AI/NIAID NIH HHS/United States
- R01AI090060/HHS | National Institutes of Health (NIH)
- K08 HL169725/HL/NHLBI NIH HHS/United States
- U54CA260582/HHS | NIH | National Cancer Institute (NCI)
- HL169725/HHS | National Institutes of Health (NIH)
- HD095881/HHS | National Institutes of Health (NIH)
- UH2 AI171611/AI/NIAID NIH HHS/United States
- R01 HD095881/HD/NICHD NIH HHS/United States
- R01 AI090060/AI/NIAID NIH HHS/United States
- P01 AI175399/AI/NIAID NIH HHS/United States
- P01AI175399/HHS | National Institutes of Health (NIH)
- T32 AI165391/AI/NIAID NIH HHS/United States
- T32AI165391/HHS | National Institutes of Health (NIH)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
