In silico construction of a multi-epitope vaccine (RGME-VAC/ATS-1) against the Rickettsia genus using immunoinformatics
- PMID: 40136144
- PMCID: PMC11932644
- DOI: 10.1590/0074-02760240201
In silico construction of a multi-epitope vaccine (RGME-VAC/ATS-1) against the Rickettsia genus using immunoinformatics
Abstract
Background: Rickettsia is a genus of Gram-negative bacteria that causes various diseases, including epidemic typhus, Rocky Mountain spotted fever, and Mediterranean spotted fever. Ticks transmit these diseases and commonly found in developing regions with poor sanitation. As a result, it is difficult to estimate the number of these diseases cases, making it challenging to create prevention and diagnostic mechanisms.
Objectives: Thus, this study aimed to develop an in silico multi-epitope vaccine against Rickettsia.
Methods: Eight proteins were previously identified as potential vaccine candidates through reverse vaccinology and were screened for epitopes that bind to MHC class I and II molecules. The epitopes were then analysed for antigenicity, allergenicity, and toxicity. The selected epitopes were linked with AAY and GPGPG sequences peptide and a known adjuvant, the B-chain of Escherichia coli heat-labile enterotoxin, to form a chimeric multi-epitope protein. The protein's three-dimensional structure was predicted, and molecular docking analysis was performed against the toll-like receptor 4 (TLR4). Finally, the immune response to the protein was simulated using C-ImmSim tool.
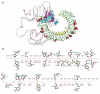
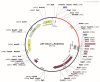
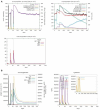
Findings: A total of 26 immunogenic epitopes, formed the multi-epitope vaccine RGME-VAC/ATS-1. The vaccine showed excellent immunogenic parameters and was predicted to do not be toxic or allergenic to the host. It also showed good potential stimulation of immune cells, with a propensity to generate memory cells and elicit IFN-γ secretion.
Main conclusions: The in silico validations suggest that our study successfully designed an innovative multi-epitope vaccine against Rickettsia, addressing the challenges posed by the elusive nature of diseases caused by this genus. We provide a promising potential for further experimental exploration and the development of targeted prevention and diagnostic strategies for these diseases.
Conflict of interest statement
All authors read and approved the final version. The authors declare that they have no competing interests with this manuscript.
Figures




References
-
- Lima DS, Farias EVS, Ferreira DA, Nascimento do LEA, Luis JAS, Lima IO. Aspectos do gênero Rickettsia uma revisão sistemática. Educ Ci e Sal. 2020;7:301–315. doi: 10.20438/ecs.v7i1.265. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

