Thonningianin A disrupts pA104R-DNA binding and inhibits African swine fever virus replication
- PMID: 40138179
- PMCID: PMC11966994
- DOI: 10.1080/22221751.2025.2482697
Thonningianin A disrupts pA104R-DNA binding and inhibits African swine fever virus replication
Abstract
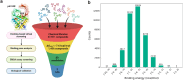
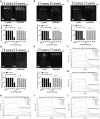
African swine fever is a highly lethal disease caused by the African swine fever virus (ASFV), posing a significant threat to the global pig industry, wherease no approved treatments are currently available. The ASFV DNA-binding protein, pA104R, plays a critical role in viral genome packaging and replication, making it a key target for drug discovery. Through structure-based virtual screening, we identified a polyphenolic compound, thonningianin A, which disrupts the pA104R-DNA binding and significantly inhibits ASFV replication. Mechanistic study revealed that thonningianin A binds to the DNA-binding region of pA104R, forming strong hydrogen bonds with H100 and occupying the vital DNA-binding residues K92, R94, and K97. In addition, we resolved the high-resolution (1.8 Å) structure of pA104R (PDB ID 9JS5), providing valuable insights for future drug screening. Together, these results demonstrate that thonningianin A holds great potential for the development of anti-ASFV drug, as a herb extract with favourable pharmacokinetic properties and safety.
Keywords: ASFV; DNA-binding protein; EMSA; pA104R; structure-based virtual screening; thonningianin A.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
