Comparison of Molnupiravir Exposure-Response Relationships for Virology Response and Mechanism of Action Biomarkers With Clinical Outcomes in Treatment of COVID-19
- PMID: 40138219
- PMCID: PMC11939005
- DOI: 10.1111/cts.70184
Comparison of Molnupiravir Exposure-Response Relationships for Virology Response and Mechanism of Action Biomarkers With Clinical Outcomes in Treatment of COVID-19
Abstract
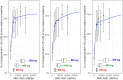
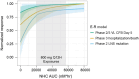
Molnupiravir, an orally administered drug for the treatment of mild-to-moderate COVID-19, is a prodrug of the ribonucleoside β-D-N4-hydroxycytidine (NHC). NHC incorporation in the SARS-CoV-2 RNA strand causes an accumulation of deleterious errors in the genome, resulting in reduced viral infectivity and replication. Exposure-response (E-R) analyses for viral RNA mutation rate and virologic outcomes were conducted using data from three phase 2/3 studies of molnupiravir (P006, MOVe-IN, and MOVe-OUT). Three dose levels (200, 400, and 800 mg every 12 hours [Q12H]) and placebo were evaluated. E-R datasets were generated for SARS-CoV-2 RNA mutation and longitudinal SARS-CoV-2 RNA viral load. E-R models were defined for RNA mutation rate and viral load change from baseline at days 5 and 10. The models supported plasma NHC AUC0-12 as the appropriate pharmacokinetic driver for assessing E-R relationships. The highest percentage of participants with > 20 low-frequency nucleotide substitutions (LNS) per 10,000 bases, a measure of likely meaningful drug effect, was predicted in the 800 mg Q12H treatment group. A strong drug effect on the reduction of viral load was observed on days 5 and 10. E-R relationships were best represented by an Emax structural model with reasonable consistency in the estimated AUC50s (~2.3-fold), across the models, of 10,260 and 4390 nM*hr. for day 5 viral load change from baseline and LNS error rate, respectively. These biomarker E-R curves support the choice of 800 mg Q12H as providing near-maximal drug effect, consistent with findings from the previously published molnupiravir E-R model of clinical outcomes.
Keywords: NHC; SARS‐CoV‐2; exposure‐response; low‐frequency nucleotide substitutions; pharmacokinetics‐pharmacodynamics; viral load; virologic outcomes; β‐D‐N4‐hydroxycytidine.
© 2025 Merck Sharp & Dohme LLC. Ridgeback Biotherapeutics and The Author(s). Clinical and Translational Science published by Wiley Periodicals LLC on behalf of American Society for Clinical Pharmacology and Therapeutics.
Conflict of interest statement
A.C., R.B., H.W., J.S., A.F., Y.C., B.M.M., A.P., C.d.A., W.G., M.L.R., and J.A.S. are/were employees of Merck Sharp & Dohme LLC, a subsidiary of Merck & Co. Inc., Rahway, NJ, USA, and may hold stock or stock options in Merck & Co. Inc., Rahway, NJ, USA. W.H. is an owner and cofounder, and advisor to Ridgeback Biotherapeutics LP, Miami, FL, USA, and is listed as an inventor on patent applications relating to molnupiravir and owns stock and/or stock options in Merck & Co. Inc., Rahway, NJ, USA. W.P. is an employee of Ridgeback Biotherapeutics. S.S. is an employee of Simulations Plus, Cognigen Division, which was contracted by Merck & Co., Inc., Rahway, NJ, USA, to perform the analysis reported here. G.P. receives royalties under the Emory license on the sale of molnupiravir, is listed as an inventor on multiple issued and pending patent applications relating to molnupiravir, and is an advisor to Ridgeback Biotherapeutics LP, Miami, Florida, USA.
Figures


References
-
- U. S. Food and Drug Administration . “Coronavirus (COVID‐19) Update: FDA Authorizes Additional Oral Antiviral for Treatment of COVID‐19 in Certain Adults,” 2021, https://www.fda.gov/news‐events/press‐announcements/coronavirus‐covid‐19....
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

