The actin and microtubule network regulator WHAMM is identified as a key kidney disease risk gene
- PMID: 40138314
- PMCID: PMC12181807
- DOI: 10.1016/j.celrep.2025.115462
The actin and microtubule network regulator WHAMM is identified as a key kidney disease risk gene
Abstract
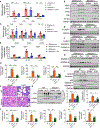
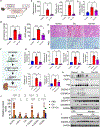
Nearly 850 million people suffer from kidney disease worldwide. Genome-wide association studies identify genetic variations at more than 800 loci associated with kidney dysfunction; however, the target genes, cell types, and mechanisms remain poorly understood. Here, we show that nucleotide variants on chromosome 15 are not only associated with kidney dysfunction but also regulate the expression of Wasp homolog associated with actin, membranes, and microtubules (WHAMM). WHAMM expression is higher in mice and patients with chronic and acute kidney disease. Mice with genetic deletion of Whamm appear healthy at baseline but develop less injury following cisplatin, folic acid, and unilateral ureteral obstruction. In vitro cell studies indicate that WHAMM controls cell death by regulating actin-mediated cytochrome c release from mitochondria and the formation of ASC speck. Pharmacological inhibition of actin dynamics mitigates kidney disease in experimental models. In summary, our study identifies a key role of WHAMM in the development of kidney disease.
Keywords: AKI; CKD; CP: Cell biology; GWAS; WASP family; WHAMM; actin cytoskeleton; autophagy; cell death; kidney tubules.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

Similar articles
-
Pharmacological and electronic cigarette interventions for smoking cessation in adults: component network meta-analyses.Cochrane Database Syst Rev. 2023 Sep 12;9(9):CD015226. doi: 10.1002/14651858.CD015226.pub2. Cochrane Database Syst Rev. 2023. PMID: 37696529 Free PMC article.
-
The genetics of low and high birthweight and their relationship with cardiometabolic disease.Diabetologia. 2025 Jul;68(7):1452-1462. doi: 10.1007/s00125-025-06420-8. Epub 2025 Apr 10. Diabetologia. 2025. PMID: 40210729 Free PMC article.
-
Assessing the comparative effects of interventions in COPD: a tutorial on network meta-analysis for clinicians.Respir Res. 2024 Dec 21;25(1):438. doi: 10.1186/s12931-024-03056-x. Respir Res. 2024. PMID: 39709425 Free PMC article. Review.
-
Salmonella exploits host- and bacterial-derived β-alanine for replication inside host macrophages.Elife. 2025 Jun 19;13:RP103714. doi: 10.7554/eLife.103714. Elife. 2025. PMID: 40536105 Free PMC article.
-
Molecular feature-based classification of retroperitoneal liposarcoma: a prospective cohort study.Elife. 2025 May 23;14:RP100887. doi: 10.7554/eLife.100887. Elife. 2025. PMID: 40407808 Free PMC article.
References
-
- Evans M, Lewis RD, Morgan AR, Whyte MB, Hanif W, Bain SC, Davies S, Dashora U, Yousef Z, Patel DC, and Strain WD (2022). A Narrative Review of Chronic Kidney Disease in Clinical Practice: Current Challenges and Future Perspectives. Adv. Ther 39, 33–43. 10.1007/s12325-021-01927-z. - DOI - PMC - PubMed
-
- Tuttle KR, Alicic RZ, Duru OK, Jones CR, Daratha KB, Nicholas SB, McPherson SM, Neumiller JJ, Bell DS, Mangione CM, and Norris KC (2019). Clinical Characteristics of and Risk Factors for Chronic Kidney Disease Among Adults and Children: An Analysis of the CURECKD Registry. JAMA Netw. Open 2, e1918169. 10.1001/jamanetworkopen.2019.18169. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

