An in vivo screen identifies NAT10 as a master regulator of brain metastasis
- PMID: 40138393
- PMCID: PMC11939035
- DOI: 10.1126/sciadv.ads6021
An in vivo screen identifies NAT10 as a master regulator of brain metastasis
Abstract
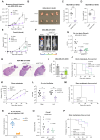
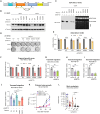
Emerging evidence has shown that epigenetic regulation plays a fundamental role in cancer metastasis, the major cause of cancer-related deaths. Here, we conducted an in vivo screen for vulnerabilities of brain metastasis and identified N-acetyltransferase 10 (NAT10) as a driver of brain metastasis. Knockdown of NAT10 restrains cancer cell proliferation and migration in vitro and tumor growth and brain metastasis in vivo. The poorly characterized RNA helicase domain of NAT10 is critical for cell growth in vitro, while both RNA helicase and NAT domains are essential for primary tumor growth and brain metastasis in vivo. Mechanically, NAT10 promotes the expression of 3-phosphoglycerate dehydrogenase (PHGDH) and phosphoserine aminotransferase 1 (PSAT1), two enzymes for serine biosynthesis implicated in brain metastasis. Silencing PHGDH or PSAT1 in metastatic breast cancer cells inhibits their growth in the serine/glycine-limited condition, phenocopying the effects of NAT10 depletion. These findings establish NAT10 as a key regulator of brain metastasis and nominate NAT10 as a target for treating metastasis.
Figures




References
-
- Martin A. M., Cagney D. N., Catalano P. J., Warren L. E., Bellon J. R., Punglia R. S., Claus E. B., Lee E. Q., Wen P. Y., Haas-Kogan D. A., Alexander B. M., Lin N. U., Aizer A. A., Brain metastases in newly diagnosed breast cancer: A population-based study. JAMA Oncol. 3, 1069–1077 (2017). - PMC - PubMed
-
- Cagney D. N., Martin A. M., Catalano P. J., Redig A. J., Lin N. U., Lee E. Q., Wen P. Y., Dunn I. F., Bi W. L., Weiss S. E., Haas-Kogan D. A., Alexander B. M., Aizer A. A., Incidence and prognosis of patients with brain metastases at diagnosis of systemic malignancy: A population-based study. Neuro Oncol. 19, 1511–1521 (2017). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

