A dynamic structural unit of phase-separated heterochromatin protein 1α as revealed by integrative structural analyses
- PMID: 40138713
- PMCID: PMC11930357
- DOI: 10.1093/nar/gkaf154
A dynamic structural unit of phase-separated heterochromatin protein 1α as revealed by integrative structural analyses
Abstract
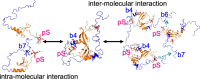
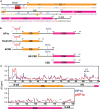
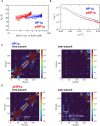
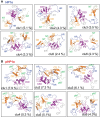
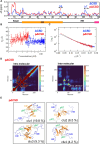
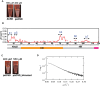
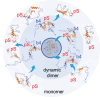
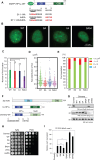
The heterochromatin protein HP1α consists of an N-terminal disordered tail (N-tail), chromodomain (CD), hinge region (HR), and C-terminal chromo shadow domain (CSD). While CD binds to the lysine9-trimethylated histone H3 (H3K9me3) tail in nucleosomes, CSD forms a dimer bridging two nucleosomes with H3K9me3. Phosphorylation of serine residues in the N-tail enhances both H3K9me3 binding and liquid-liquid phase separation (LLPS) by HP1α. We have used integrative structural methods, including nuclear magnetic resonance, small-angle X-ray scattering (SAXS), and multi-angle-light scattering combined with size-exclusion chromatography, and coarse-grained molecular dynamics simulation with SAXS, to probe the HP1α dimer and its CSD deletion monomer. We show that dynamic intra- and intermolecular interactions between the N-tails and basic segments in CD and HR depend on N-tail phosphorylation. While the phosphorylated HP1α dimer undergoes LLPS via the formation of aggregated multimers, the N-tail phosphorylated mutant without CSD still undergoes LLPS, but its structural unit is a dynamic intermolecular dimer formed via the phosphorylated N-tail and a basic segment at the CD end. Furthermore, we reveal that mutation of this basic segment in HP1α affects the size of heterochromatin foci in cultured mammalian cells, suggesting that this interaction plays an important role in heterochromatin formation in vivo.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

