Evidence supporting a catalytic pentad mechanism for the proteasome and other N-terminal nucleophile enzymes
- PMID: 40140419
- PMCID: PMC11947121
- DOI: 10.1038/s41467-025-58077-x
Evidence supporting a catalytic pentad mechanism for the proteasome and other N-terminal nucleophile enzymes
Abstract
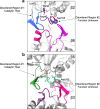
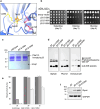
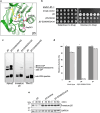
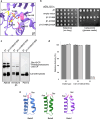
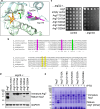
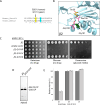
Proteases are defined by their nucleophile but require additional residues to regulate their active sites, most often arranged as catalytic triads that control the generation and resolution of acyl-enzyme intermediates. Threonine N-terminal nucleophiles represent a diverse family of proteases and transferases that possess two active site nucleophiles, the side chain hydroxyl and the free amino-terminus, and require autocatalytic cleavage of their N-terminal propeptides. Here we provide evidence that the proteasome, which mediates intracellular protein degradation and contains three different threonine protease subunits, utilizes a unique catalytic pentad mechanism. In addition to the previously defined lysine/aspartate pair which regulates threonine's side chain, a second serine/aspartate pair appears to regulate threonine's amino-terminus. The pentad is required for substrate proteolysis and assembly-coupled autocatalytic cleavage, the latter triggered by alignment of the full pentad upon fusion of two half-proteasome precursors. A similar pentad mechanism was required by the ornithine acetyltransferase Arg7, suggesting that this may be a general property of threonine N-terminal nucleophiles. Finally, we show that two patient-derived proteasome mutations compromise function of the serine/aspartate unit in yeast, suggesting that defective pentad function may underlie some human proteasomopathies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Mechanism of autocatalytic activation during proteasome assembly.Nat Struct Mol Biol. 2024 Aug;31(8):1167-1175. doi: 10.1038/s41594-024-01262-1. Epub 2024 Apr 10. Nat Struct Mol Biol. 2024. PMID: 38600323 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Toward pharmacologic therapy for glioblastoma: Identifying inhibitors of very long-chain acyl-CoA synthetase 3 (ACSVL3).bioRxiv [Preprint]. 2025 Jul 3:2025.07.02.662811. doi: 10.1101/2025.07.02.662811. bioRxiv. 2025. PMID: 40631213 Free PMC article. Preprint.
-
The ribonuclease RNase T2 mediates selective autophagy of ribosomes induced by starvation in Saccharomyces cerevisiae.J Biol Chem. 2025 Jun;301(6):108554. doi: 10.1016/j.jbc.2025.108554. Epub 2025 Apr 26. J Biol Chem. 2025. PMID: 40294649 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
References
-
- Brannigan, J. A. et al. A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature378, 416–419 (1995). - PubMed
-
- Seemuller, E. et al. Proteasome from thermoplasma acidophilum: a threonine protease. Science268, 579–582 (1995). - PubMed
-
- Seemuller, E., Lupas, A. & Baumeister, W. Autocatalytic processing of the 20S proteasome. Nature382, 468–471 (1996). - PubMed
-
- Groll, M. et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature386, 463–471 (1997). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

