The histone modifier KAT2A presents a selective target in a subset of well-differentiated microsatellite-stable colorectal cancers
- PMID: 40140561
- PMCID: PMC12284170
- DOI: 10.1038/s41418-025-01479-7
The histone modifier KAT2A presents a selective target in a subset of well-differentiated microsatellite-stable colorectal cancers
Abstract
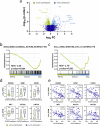
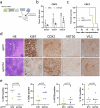
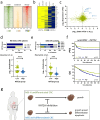
Lysine acetyltransferase 2 A (KAT2A) plays a pivotal role in epigenetic gene regulation across various types of cancer. In colorectal cancer (CRC), increased KAT2A expression is associated with a more aggressive phenotype. Our study aims to elucidate the molecular underpinnings of KAT2A dependency in CRC and assess the consequences of KAT2A depletion. We conducted a comprehensive analysis by integrating CRISPR-Cas9 screening data with genomics, transcriptomics, and global acetylation patterns in CRC cell lines to pinpoint molecular markers indicative of KAT2A dependency. Additionally, we characterized the phenotypic effect of a CRISPR-interference-mediated KAT2A knockdown in CRC cell lines and patient-derived 3D spheroid cultures. Moreover, we assessed the effect of KAT2A depletion within a patient-derived xenograft mouse model in vivo. Our findings reveal that KAT2A dependency is closely associated with microsatellite stability, lower mutational burden, and increased molecular differentiation signatures in CRC, independent of the KAT2A expression levels. KAT2A-dependent CRC cells display higher gene expression levels and enriched H3K27ac marks at gene loci linked to enterocytic differentiation. Furthermore, loss of KAT2A leads to decreased cell growth and viability in vitro and in vivo, downregulation of proliferation- and stem cell-associated genes, and induction of differentiation markers. Altogether, our data show that a specific subset of CRCs with a more differentiated phenotype relies on KAT2A. For these CRC cases, KAT2A might represent a promising novel therapeutic target.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no conflict of interest.
Figures



References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. - PubMed
-
- Dieter SM, Siegl C, Codó PL, Huerta M, Ostermann-Parucha AL, Schulz E, et al. Degradation of CCNK/CDK12 is a druggable vulnerability of colorectal cancer. Cell Rep. 2021;36:109394. - PubMed
-
- Weichert W, Röske A, Gekeler V, Beckers T, Ebert MP, Pross M, et al. Association of patterns of class I histone deacetylase expression with patient prognosis in gastric cancer: a retrospective analysis. Lancet Oncol. 2008;9:139–48. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

