BRCA2 prevents PARPi-mediated PARP1 retention to protect RAD51 filaments
- PMID: 40140565
- PMCID: PMC12161360
- DOI: 10.1038/s41586-025-08749-x
BRCA2 prevents PARPi-mediated PARP1 retention to protect RAD51 filaments
Abstract
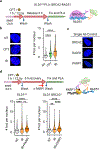
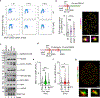
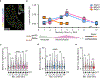
The tumour-suppressor protein BRCA2 has a central role in homology-directed DNA repair by enhancing the formation of RAD51 filaments on resected single-stranded DNA generated at double-stranded DNA breaks and stimulating RAD51 activity1,2. Individuals with BRCA2 mutations are predisposed to cancer; however, BRCA2-deficient tumours are often responsive to targeted therapy with PARP inhibitors (PARPi)3-6. The mechanism by which BRCA2 deficiency renders cells sensitive to PARPi but with minimal toxicity in cells heterozygous for BRCA2 mutations remains unclear. Here we identify a previously unknown role of BRCA2 that is directly linked to the effect of PARP1 inhibition. Using biochemical and single-molecule approaches, we demonstrate that PARPi-mediated PARP1 retention on a resected DNA substrate interferes with RAD51 filament stability and impairs RAD51-mediated DNA strand exchange. Full-length BRCA2 protects RAD51 filaments and counteracts the instability conferred by PARPi-mediated retention by preventing the binding of PARP1 to DNA. Extending these findings to a cellular context, we use quantitative single-molecule localization microscopy to show that BRCA2 prevents PARPi-induced PARP1 retention at homologous-recombination repair sites. By contrast, BRCA2-deficient cells exhibit increased PARP1 retention at these lesions in response to PARPi. These results provide mechanistic insights into the role of BRCA2 in maintaining RAD51 stability and protecting homologous-recombination repair sites by mitigating PARPi-mediated PARP1 retention.
© 2025. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests: R.B.J. is named on patent US9150897B2, which references the phCMV1-2XMBP vector used to purify full-length BRCA2 and PARP1 used in this study. The other authors declare no competing interests.
Figures











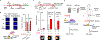
References
-
- McCabe N et al. BRCA2-deficient CAPAN-1 cells are extremely sensitive to the inhibition of Poly (ADP-Ribose) polymerase: an issue of potency. Cancer Biol Ther 4, 934–936 (2005). - PubMed
-
- Farmer H et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005). - PubMed
-
- Fong PC et al. Inhibition of Poly(ADP-Ribose) Polymerase in Tumors from BRCA Mutation Carriers. New Engl J Med 361, 123–134 (2009). - PubMed
Associated References
-
- Jensen R Purification of recombinant 2XMBP tagged human proteins from human cells. Methods Mol Biol 1176, 209–217 (2014). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

