The contribution of de novo coding mutations to meningomyelocele
- PMID: 40140573
- PMCID: PMC12625549
- DOI: 10.1038/s41586-025-08676-x
The contribution of de novo coding mutations to meningomyelocele
Abstract
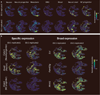
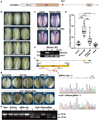
Meningomyelocele (also known as spina bifida) is considered to be a genetically complex disease resulting from a failure of the neural tube to close. Individuals with meningomyelocele display neuromotor disability and frequent hydrocephalus, requiring ventricular shunting. A few genes have been proposed to contribute to disease susceptibility, but beyond that it remains unexplained1. We postulated that de novo mutations under purifying selection contribute to the risk of developing meningomyelocele2. Here we recruited a cohort of 851 meningomyelocele trios who required shunting at birth and 732 control trios, and found that de novo likely gene disruption or damaging missense mutations occurred in approximately 22.3% of subjects, with 28% of such variants estimated to contribute to disease risk. The 187 genes with damaging de novo mutations collectively define networks including actin cytoskeleton and microtubule-based processes, Netrin-1 signalling and chromatin-modifying enzymes. Gene validation demonstrated partial or complete loss of function, impaired signalling and defective closure of the neural tube in Xenopus embryos. Our results indicate that de novo mutations make key contributions to meningomyelocele risk, and highlight critical pathways required for neural tube closure in human embryogenesis.
© 2025. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests: A. Alkelai and A.R.S. are full-time employees of Regeneron Genetics Center. S.K. is a cofounder of AIMA, which seeks to develop techniques for early cancer diagnosis based on circulating tumour DNA. R.H.F. previously led TeratOmic Consulting, which is now defunct, and received travel funds for Reproductive and Developmental Medicine editorial board meetings.
Figures
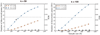







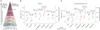



References
-
- Iskandar BJ & Finnell RH Spina bifida. N. Engl. J. Med 387, 444–450 (2022). - PubMed
-
- MRC Vitamin Study Research Group. Prevention of neural tube defects: results of the Medical Research Council vitamin study. Lancet 338, 131–137 (1991). - PubMed
-
- Arnold JA Myelocyste, transposition von gewebskeimen und sympodie. Beitr. Pathol. Anat 16, 1–28 (1894).
-
- Chiari H. Uber veränderungen des kleinhirns infolge von hydrocephalie des grosshirns. Dtsch. Med. Wochenschr 17, 1172–1175 (1891).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

