Socio-economic status is a social construct with heritable components and genetic consequences
- PMID: 40140606
- PMCID: PMC7617559
- DOI: 10.1038/s41562-025-02150-4
Socio-economic status is a social construct with heritable components and genetic consequences
Abstract
In civilizations, individuals are born into or sorted into different levels of socio-economic status (SES). SES clusters in families and geographically, and is robustly associated with genetic effects. Here we first review the history of scientific research on the relationship between SES and heredity. We then discuss recent findings in genomics research in light of the hypothesis that SES is a dynamic social construct that involves genetically influenced traits that help in achieving or retaining a socio-economic position, and can affect the distribution of genes associated with such traits. Social stratification results in people with differing traits being sorted into strata with different environmental exposures, which can result in evolutionary selection pressures through differences in mortality, reproduction and non-random mating. Genomics research is revealing previously concealed genetic consequences of the way society is organized, yielding insights that should be approached with caution in pursuit of a fair and functional society.
© 2025. Springer Nature Limited.
Conflict of interest statement
Competing interests: M.C.M. is a trustee of the UK Biobank, is on the Scientific Advisory Board of Our Future Health and Lifelines Biobank, and is on the Data Management Advisory Board of the Health and Retirement Survey. F.C.T. is a research fellow at AnalytiXIN, which is a consortium of health-data organizations, industry partners and university partners in Indiana primarily funded through the Lilly Endowment, IU Health and Eli Lilly and Company. The remaining authors declare no competing interests.
Figures
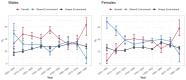


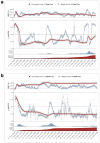
References
-
- Abdellaoui A, et al. Genetic correlates of social stratification in Great Britain. Nature human behaviour. 2019;3:1332–1342. - PubMed
-
- Flannery K, Marcus J. The creation of inequality: how our prehistoric ancestors set the stage for monarchy, slavery, and empire. Harvard University Press; 2012.
-
- Alesina A, La Ferrara E. Participation in heterogeneous communities. The quarterly journal of economics. 2000;115:847–904.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

