C9ORF72 poly-PR disrupts expression of ALS/FTD-implicated STMN2 through SRSF7
- PMID: 40140908
- PMCID: PMC11948778
- DOI: 10.1186/s40478-025-01977-2
C9ORF72 poly-PR disrupts expression of ALS/FTD-implicated STMN2 through SRSF7
Abstract
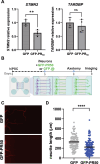
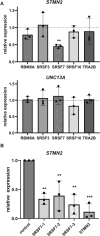
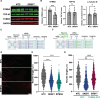
A hexanucleotide repeat expansion in C9ORF72 is the most common genetic cause of amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD), and combined ALS/FTD. The repeat is transcribed in the sense and the antisense directions to produce several dipeptide repeat proteins (DPRs) that have toxic gain-of-function effects; however, the mechanisms by which DPRs lead to neural dysfunction remain unresolved. Here, we observed that poly-proline-arginine (poly-PR) was sufficient to inhibit axonal regeneration of human induced pluripotent stem cell (iPSC)-derived neurons. Global phospho-proteomics revealed that poly-PR selectively perturbs nuclear RNA binding proteins (RBPs). In neurons, we found that depletion of one of these RBPs, SRSF7 (serine/arginine-rich splicing factor 7), resulted in decreased abundance of STMN2 (stathmin-2), though not TDP-43. STMN2 supports axon maintenance and repair and has been recently implicated in the pathogenesis of ALS/FTD. We observed that depletion of SRSF7 impaired axonal regeneration, a phenotype that could be rescued by exogenous STMN2. We propose that antisense repeat-encoded poly-PR perturbs RBPs, particularly SRSF7, resulting in reduced STMN2 and axonal repair defects in neurons. Hence, we provide a potential link between DPRs gain-of-function effects and STMN2 loss-of-function phenotypes in neurodegeneration.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The usage of human induced pluripotent stem cells (iPSCs) was approved by the Human Gamete, Embryo and Stem Cell Research (GESCR) Committee at UCSF. Consent for publication: Not applicable. Competing interests: K.E. is a co-founder of Quralis and Enclear Therapies, and currently employed at BioMarin Pharmaceutical. D.A.M. receives sponsored research support from Genentech/Roche that is unrelated to this study. The remaining authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

