Complementary role of transcriptomic endotyping and protein-based biomarkers for risk stratification in sepsis-associated acute kidney injury
- PMID: 40140945
- PMCID: PMC11948859
- DOI: 10.1186/s13054-025-05361-3
Complementary role of transcriptomic endotyping and protein-based biomarkers for risk stratification in sepsis-associated acute kidney injury
Abstract
Background: Sepsis-associated acute kidney injury (SA-AKI) is a prevalent and severe complication in critically ill patients. However, diagnostic and therapeutic advancements have been hindered by the biological heterogeneity underlying the disease. Both transcriptomic endotyping and biomarker profiling have been proposed individually to identify molecular subtypes of sepsis and may enhance risk stratification. This study aimed to evaluate the utility of combining transcriptomic endotyping with protein-based biomarkers for improving risk stratification in SA-AKI.
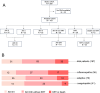
Methods: This secondary analysis of the PredARRT-Sep-Trial included 167 critically ill patients who met Sepsis-3 criteria. Patients were stratified into three transcriptomic endotypes-inflammopathic (IE), adaptive (AE), and coagulopathic (CE)-using a validated whole-blood gene expression classifier. Eight protein-based biomarkers encompassing kidney function, vascular integrity, and immune response were measured. Predictive performance for the primary endpoint kidney replacement therapy or death was assessed using receiver operating characteristic curve analysis and logistic regression models.
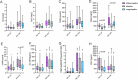
Results: Stratification into transcriptomic endotypes assigned 33% of patients to IE, 42% to AE, and 24% to CE. Patients classified as IE exhibited the highest disease severity and were most likely to meet the primary endpoint (30%), compared to AE and CE (17% and 10%, respectively). Kidney function biomarkers showed stepwise increases with AKI severity across all endotypes, whereas non-functional biomarkers (neutrophil gelatinase-associated lipocalin [NGAL], soluble urokinase plasminogen activator receptor [suPAR], and bioactive adrenomedullin [bio-ADM]) exhibited endotype-specific differences independent of AKI severity. NGAL and suPAR levels were disproportionately elevated in the IE group, suggesting a dominant role of innate immune dysregulation in this endotype. In contrast, bio-ADM, a marker of endothelial dysfunction, was the strongest risk-predictor of outcomes in CE. The combination of transcriptomic endotyping with protein-based biomarkers enhanced predictive accuracy for the primary endpoint and 7-day mortality, with the highest area under the receiver operating characteristic curve of 0.80 (95% CI 0.72-0.88) for endotyping + bio-ADM and 0.85 (95% CI 0.78-0.93) for endotyping and suPAR, respectively. Combinations of endotyping with functional and non-functional biomarkers particularly improved mortality-related risk stratification.
Conclusions: Combining transcriptomic endotyping with protein-based biomarker profiling enhances risk-stratification in SA-AKI, offering a promising strategy for personalized treatment and trial enrichment in the future. Further research should validate these findings and explore therapeutic applications.
Keywords: Acute kidney injury; Biomarkers; Risk stratification; Sepsis; Transcriptomic endotyping.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study design was approved by the local Ethics Committee of the Medical Faculty of Heidelberg (S-200/2017) and registered at the German Clinical Trials Register (DRKS-ID: DRKS00012446). Written informed consent was obtained from all participants or their legal representatives. Consent for publication: Not applicable. Competing interests: Christian Nusshag received travel expense reimbursements from SphingoTec. Florian Uhle is an employee and stock option holder of Sphingotec and Inflammatix. Oliver Liesenfeld is stock option holder of Inflammatix. Tim Sweeney is an employee and stock option holder of Inflammatix. All other authors of this manuscript have no conflicts of interest to disclose as described by the journal.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

