Novel Phenotypical and Biochemical Findings in Mucolipidosis Type II
- PMID: 40141052
- PMCID: PMC11941985
- DOI: 10.3390/ijms26062408
Novel Phenotypical and Biochemical Findings in Mucolipidosis Type II
Abstract
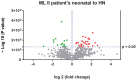
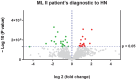
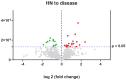
Mucolipidosis type II is a very rare lysosomal disease affecting the UDP-GlcNAc N-acetylglucosamine-1-phosphotransferase enzyme, which catalyzes the synthesis of the targeting signal mannose 6-phosphate in lysosomal acid hydrolases. Its deficiency hinders the arrival of lysosomal enzymes to the lysosome, diminishing the multiple degradations of components that cells need to perform. Due to the low prevalence of this condition, available information is scarce. This article aims to deepen the understanding of the disease; clinical, biochemical, and proteomic data are analyzed. Three patients have been identified presenting GNPTAB pathogenic variants using whole exome sequencing. A biochemical profile for these patients has been carried out through quantification of glycosaminoglycans in urine samples and enzymatic analysis in dried blood spot (DBS) samples. Quantitative proteomic studies were performed. Results show how enzymatic assays in DBS can be used to diagnose this disease both during the neonatal period or in patients of more advanced age. Increased levels of acid sphingomyelinase, alpha-iduronidase, iduronidate 2-sulfatase, alpha-N-acetyl glucosaminidase, and beta-glucuronidase are found. Conclusion: this biochemical method could potentially improve early diagnosis. Proteomic data supporting these results reveal disrupted biochemical pathways, including the degradation of dermatan sulfate, heparan sulfate, and cellular cholesterol trafficking.
Keywords: biochemistry; biomarkers; early findings; lysosomal diseases; mucolipidosis II-III; proteomic studies.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

