Deciphering the Genetic Basis of Degenerative and Developmental Eye Disorders in 50 Pakistani Consanguineous Families Using Whole-Exome Sequencing
- PMID: 40141357
- PMCID: PMC11942243
- DOI: 10.3390/ijms26062715
Deciphering the Genetic Basis of Degenerative and Developmental Eye Disorders in 50 Pakistani Consanguineous Families Using Whole-Exome Sequencing
Abstract
Degenerative and developmental eye disorders, including inherited retinal dystrophies (IRDs), anophthalmia, and congenital cataracts arise from genetic mutations, causing progressive vision loss or congenital structural abnormalities. IRDs include a group of rare, genetically, and clinically heterogeneous retinal diseases. It is caused by variations in at least 324 genes, affecting numerous retinal regions. In addition to IRDs, other developmental eye disorders such as anophthalmia and congenital cataracts also have a strong genetic basis. Autosomal recessive IRDs, anophthalmia, and congenital cataracts are common in consanguineous populations. In many endogamous populations, including those in Pakistan, a significant proportion of IRD and anophthalmia cases remain genetically undiagnosed. The present study investigated the variations in IRDs, anophthalmia, and congenital cataracts genes in 50 affected families. These unrelated consanguineous families were recruited from the different provinces of Pakistan including Punjab, Khyber Pakhtoon Khwa, Sindh, Gilgit Baltistan, and Azad Kashmir. Whole exome sequencing (WES) was conducted for the proband of each family. An in-house customized pipeline examined the data, and bioinformatics analysis predicted the pathogenic effects of identified variants. The relevant identified DNA variants of selected families were assessed in parents and healthy siblings via Sanger sequencing. WES identified 12 novel variants across 10 known IRD-associated genes. The four most frequently implicated genes were CRB1 (14.3%), GUCY2D (9.5%), AIPL1 (9.5%), and CERKL (7.1%) that together accounted for 40.4% of all molecularly diagnosed cases. Additionally, 25 reported variants in 19 known IRDs, anophthalmia, and congenital cataracts-associated genes were found. Among the identified variants, p. Trp278X, a stop-gain mutation in the AIPL1 (NM_014336) gene, was the most common causative variant detected. The most frequently observed phenotype was retinitis pigmentosa (46.5%) followed by Leber congenital amaurosis (18.6%). Furthermore, 98% of pedigrees (49 out of 50) were affected by autosomal recessive IRDs, anophthalmia and congenital cataracts. The discovery of 12 novel likely pathogenic variants in 10 IRD genes, 25 reported variants in 19 known IRDs, anophthalmia and congenital cataracts genes, atypical phenotypes, and inter and intra-familial variability underscores the genetic and phenotypic heterogeneity of developmental and degenerative eye disorders in the Pakistani population and further expands the mutational spectrum of genes associated with these ocular disorders.
Keywords: IRDs; Pakistani population; WES; anophthalmia; autosomal recessive; degenerative and developmental eye disorder; genetic analysis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
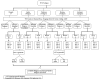
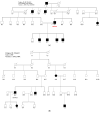
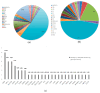


Similar articles
-
Deciphering the genetic architecture and ethnographic distribution of IRD in three ethnic populations by whole genome sequence analysis.PLoS Genet. 2021 Oct 18;17(10):e1009848. doi: 10.1371/journal.pgen.1009848. eCollection 2021 Oct. PLoS Genet. 2021. PMID: 34662339 Free PMC article.
-
Phenotypic and Genetic Heterogeneity of a Pakistani Cohort of 15 Consanguineous Families Segregating Variants in Leber Congenital Amaurosis-Associated Genes.Genes (Basel). 2024 Dec 21;15(12):1646. doi: 10.3390/genes15121646. Genes (Basel). 2024. PMID: 39766915 Free PMC article.
-
Next-generation sequencing to genetically diagnose a diverse range of inherited eye disorders in 15 consanguineous families from Pakistan.Exp Eye Res. 2024 Jul;244:109945. doi: 10.1016/j.exer.2024.109945. Epub 2024 May 28. Exp Eye Res. 2024. PMID: 38815792
-
A systematic review of inherited retinal dystrophies in Pakistan: updates from 1999 to April 2023.BMC Ophthalmol. 2024 Feb 5;24(1):55. doi: 10.1186/s12886-024-03319-7. BMC Ophthalmol. 2024. PMID: 38317096 Free PMC article.
-
State of the Art on Inherited Retinal Dystrophies: Management and Molecular Genetics.J Clin Med. 2025 May 18;14(10):3526. doi: 10.3390/jcm14103526. J Clin Med. 2025. PMID: 40429522 Free PMC article. Review.
References
-
- Tavares J., Lorenz B., Born I., Marques J., Stingl K., Pilotto E., Issa P., Leroux D., Dollfus H., Scholl H. Current management of Inherited Retinal Degenerations (IRD) patients in Europe. Results of a 2 years follow-up multinational survey by EVICR. net and ERN-EYE. Investig. Ophthalmol. Vis. Sci. 2022;63:4481-F0268.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

