Pulmonary Myeloid Cells in Mild Cases of COVID-19 Upregulate the Intracellular Fc Receptor TRIM21 and Transcribe Proteasome-Associated Molecules
- PMID: 40141410
- PMCID: PMC11943277
- DOI: 10.3390/ijms26062769
Pulmonary Myeloid Cells in Mild Cases of COVID-19 Upregulate the Intracellular Fc Receptor TRIM21 and Transcribe Proteasome-Associated Molecules
Abstract
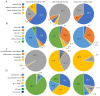
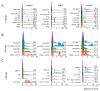
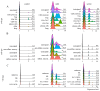
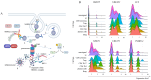
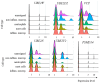
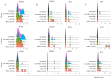
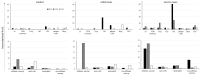
Much remains to be understood about COVID-19, but the protective role of antibodies (Igs) is widely accepted in SARS-CoV-2 infection. Igs' functions are mainly carried out by receptors that bind to their Fc portion (FcR), and less attention has been dedicated to the cytoplasmic members of this family. In this work, we used single-cell RNA sequencing (scRNA-seq) data to discern cell populations in bronchoalveolar lavage fluid obtained from healthy individuals and patients with mild or severe COVID-19. Then, we evaluated the transcription of neonatal FcR (FcRn, FCGRT gene) and tripartite motif-containing protein 21 (TRIM21) and its downstream signaling components. The TRIM21 pathway is vital for virus infections as it has a dual function, leading opsonized viruses to degradation by proteasomes and the activation of innate inflammatory anti-virus response. The transcriptional level of FCGRT showed no statistical differences in any cell population comparing the three groups of patients. On the other hand, TRIM21 transcription was significantly higher in myeloid cells collected from patients with mild COVID-19. When comparing mild with severe cases, there was no statistical difference in TRIM21 transcription in lung adaptive lymphoid cells and innate lymphoid cells (ILC). Yet, we analyzed the transcription of all downstream signaling molecules in myeloid and, as most cells expressed the receptor, in adaptive lymphoid cells. Moreover, ILCs from mild cases and all cell populations from severe cases were missing most downstream components of the pathway. We observed that members of the ubiquitin-proteasome system (UPS) and other components associated with TRIM21 proteasomal degradation were transcribed in mild cases. Despite the transcription of the danger sensors DDX58 and IFIH1, the transcriptional level of inflammatory IL1B and IL18 was generally very low, along with the NLRP3 danger sensor, members of the NF-κB pathway, and TNF. Therefore, our data suggest that TRIM21 may contribute to SARS-CoV-2 protection by reducing the viral load, while the inflammatory branch of the pathway would be silenced, leading to no pathogenic cytokine production.
Keywords: COVID-19; Fc receptors; TRIM21; inflammatory response; scRNA-seq.
Conflict of interest statement
The authors declare that the research was conducted without any commercial or financial relationships that could be construed as potential conflicts of interest.
Figures


Similar articles
-
Cytosolic Internalization of Anti-DNA Antibodies by Human Monocytes Induces Production of Pro-inflammatory Cytokines Independently of the Tripartite Motif-Containing 21 (TRIM21)-Mediated Pathway.Front Immunol. 2018 Sep 4;9:2019. doi: 10.3389/fimmu.2018.02019. eCollection 2018. Front Immunol. 2018. PMID: 30233598 Free PMC article.
-
Cytosolic Fc receptor TRIM21 inhibits seeded tau aggregation.Proc Natl Acad Sci U S A. 2017 Jan 17;114(3):574-579. doi: 10.1073/pnas.1607215114. Epub 2017 Jan 3. Proc Natl Acad Sci U S A. 2017. PMID: 28049840 Free PMC article.
-
Gene disruption study reveals a nonredundant role for TRIM21/Ro52 in NF-kappaB-dependent cytokine expression in fibroblasts.J Immunol. 2009 Jun 15;182(12):7527-38. doi: 10.4049/jimmunol.0804121. J Immunol. 2009. PMID: 19494276 Free PMC article.
-
TRIM21-From Intracellular Immunity to Therapy.Front Immunol. 2019 Aug 28;10:2049. doi: 10.3389/fimmu.2019.02049. eCollection 2019. Front Immunol. 2019. PMID: 31555278 Free PMC article. Review.
-
TRIM21 and the Function of Antibodies inside Cells.Trends Immunol. 2017 Dec;38(12):916-926. doi: 10.1016/j.it.2017.07.005. Epub 2017 Aug 11. Trends Immunol. 2017. PMID: 28807517 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

