The Infectivity and Pathogenicity Characteristics of a Recombinant Porcine Epidemic Diarrhea Virus, CHFJFQ
- PMID: 40143328
- PMCID: PMC11945473
- DOI: 10.3390/v17030401
The Infectivity and Pathogenicity Characteristics of a Recombinant Porcine Epidemic Diarrhea Virus, CHFJFQ
Abstract
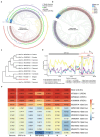
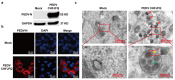
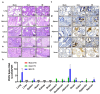
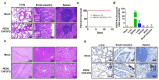
Porcine epidemic diarrhea virus (PEDV) presents a substantial challenge to the global swine industry. However, the origin, host range, and potential cross-species transmission of PEDV remain poorly understood. This study characterizes a novel PEDV strain, CHFJFQ, isolated from diarrheic piglets in Fuqing, Fujian, China. Through sequencing and phylogenetic analysis, we determined that CHFJFQ belongs to the GIIa subgroup and is a recombinant with CH/HNXX/2016 as the major parent and NW17 as the minor parent. Compared to CV777, CHFJFQ exhibits multiple base deletions and insertions across the 5'UTR, ORF1a/b, S, and ORF3 genes. Phylogenetic analysis indicates shared ancestry with bat coronaviruses, though a direct zoonotic origin remains uncertain. Interestingly, CHFJFQ demonstrated its ability to infect human and mouse cell lines in vitro and, more significantly, caused in vivo infection in both pigs and mice. The primary target organs were the intestines, lungs, and spleen, resulting in 100% mortality in suckling piglets. PEDV CHFJFQ was detected in mouse tissues, but no clinical signs were observed, indicating limited cross-species pathogenicity. Overall, these findings offer crucial insights into the epidemiology, genetics, infectivity, and pathogenicity of PEDV and provide valuable information for vaccine development.
Keywords: PEDV; cross-species transmission; infection; origin.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

