Genomic Insights into Neglected Orthobunyaviruses: Molecular Characterization and Phylogenetic Analysis
- PMID: 40143333
- PMCID: PMC11945402
- DOI: 10.3390/v17030406
Genomic Insights into Neglected Orthobunyaviruses: Molecular Characterization and Phylogenetic Analysis
Abstract
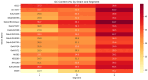
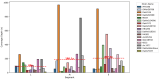
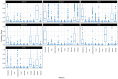
Orthobunyaviruses are a diverse group of segmented RNA viruses with significant but underexplored public and veterinary health implications. This study provides a genomic, phylogenetic, and ecological analysis of neglected Orthobunyaviruses using next-generation sequencing and computational predictions. We identified unique phylogenetic relationships, with Tanga virus forming a distinct lineage linked to zoonotic, human-associated, or non-vertebrate viruses across segments. GC content analysis revealed segment-specific patterns: higher GC content in the S segment suggests genomic stability and immune evasion, while lower GC content in the L segment reflects host-vector adaptation. Phylogenetic ties to well-characterized pathogenic viruses, such as Ilesha virus with Cache Valley virus and Bwamba virus with California encephalitis virus, indicate potential neurotropism. Ingwavuma virus clustered with Oropouche virus, suggesting risks of systemic febrile illnesses. Within the Simbu serogroup, Sango and Sabo viruses show teratogenic risks to livestock. Vector and host predictions implicate rodents, artiodactyls, and primates in Orthobunyavirus transmission, emphasizing complex ecological dynamics and zoonotic potential. These findings advance the understanding of Orthobunyavirus diversity, linking genomic features to pathogenicity and ecological adaptation, while providing a foundation for future surveillance and intervention strategies targeting these neglected viruses.
Keywords: genomic characterization; neglected Orthobunyaviruses; phylogenetics; vector-borne diseases; zoonotic potential.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of this study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


References
-
- International Committee on Taxonomy of Viruses ICTV. [En ligne]. Disponible sur. [(accessed on 9 December 2024)]. Available online: https://ictv.global/taxonomy.
-
- Toure C.T., Dieng I., Sankhe S., Kane M., Dia M., Mhamadi M., Ndiaye M., Faye O., Sall A.A., Diagne M.M., et al. Genomic Characterization of a Bataï Orthobunyavirus, Previously Classified as Ilesha Virus, from Field-Caught Mosquitoes in Senegal, Bandia 1969. Viruses. 2024;16:261. doi: 10.3390/v16020261. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

