Two Avastrovirus Species Discovered in Psittaciformes Expand the Host Range of the Family Astroviridae
- PMID: 40143376
- PMCID: PMC11946394
- DOI: 10.3390/v17030450
Two Avastrovirus Species Discovered in Psittaciformes Expand the Host Range of the Family Astroviridae
Abstract
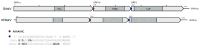
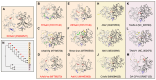
Metatranscriptomics has recently revealed greater species richness and host range of the Avastrovirus genus, quadrupling the number of avian orders known to host them in less than a decade. Despite this growing awareness of astrovirus presence in wild birds, limited attention has been paid to these viruses in the context of disease in Australian avifauna. Here we used unbiased RNA sequencing of intestinal samples from a galah (Eolophus roseicapilla) and an Australian king parrot (Alisterus scapularis) with a chronic diarrhoeal and wasting disease to detect the entire genomes of two novel astrovirus species. We propose naming these viruses Avastrovirus eolorosei (PQ893528) and Avastrovirus aliscap (PQ893527). The phylogenetic positions of these viruses highlight the importance of current and future metatranscriptomic virus screening in investigations of avian host landscapes beyond Galloanserae. This is also the first documentation of avastrovirus infections in Psittaciformes and the first to report their potential role as disease agents in them.
Keywords: Psittaciformes; astrovirus; metatranscriptomics; parrots; virus discovery.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
First Isolation and Genetic Characterization of Avian Nephritis Virus 4 from Commercial Poultry in India.Avian Dis. 2024 Sep;68(3):202-208. doi: 10.1637/aviandiseases-D-23-00053. Avian Dis. 2024. PMID: 39400214
-
Genetic diversity of astroviruses detected in wild aquatic birds in Hong Kong.Virol J. 2024 Jul 7;21(1):153. doi: 10.1186/s12985-024-02423-w. Virol J. 2024. PMID: 38972989 Free PMC article.
-
Adenosine deaminase promotes goose astrovirus genotype II replication in GEF cells.Poult Sci. 2025 Jul;104(7):105176. doi: 10.1016/j.psj.2025.105176. Epub 2025 Apr 17. Poult Sci. 2025. PMID: 40305933 Free PMC article.
-
Intra- and Cross-Species Transmission of Astroviruses.Viruses. 2021 Jun 11;13(6):1127. doi: 10.3390/v13061127. Viruses. 2021. PMID: 34208242 Free PMC article. Review.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
-
- Walker P.J., Siddell S.G., Lefkowitz E.J., Mushegian A.R., Adriaenssens E.M., Alfenas-Zerbini P., Dempsey D.M., Dutilh B.E., García M.L., Curtis Hendrickson R., et al. Recent changes to virus taxonomy ratified by the International Committee on Taxonomy of Viruses. Arch. Virol. 2022;167:2429–2440. - PMC - PubMed
-
- Wille M., Shi M., Hurt A.C., Klaassen M., Holmes E.C. RNA virome abundance and diversity is associated with host age in a bird species. Virology. 2021;561:98–106. - PubMed
Publication types
MeSH terms
Associated data
- SRA/SRR31990207-8
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

