A modern heart transplant rejection surveillance protocol utilizing cell-free DNA: A single-center experience
- PMID: 40144235
- PMCID: PMC11935408
- DOI: 10.1016/j.jhlto.2024.100076
A modern heart transplant rejection surveillance protocol utilizing cell-free DNA: A single-center experience
Abstract
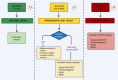
Background: Endomyocardial biopsy (EMBx) is considered the gold standard for rejection monitoring after heart transplantation; however, it is invasive and histologic interpretation has limitations. Sensitive blood biomarkers, including donor-derived cell-free DNA (dd-cfDNA), have emerged to decrease EMBx frequency.
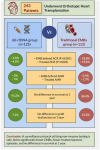
Methods: We retrospectively reviewed data on 237 patients who underwent heart transplantation at our institution. Of these, 125 patients underwent monitoring using dd-cfDNA, combined with a fewer number of EMBx, and 112 patients underwent monitoring using EMBx only. We compared rates of rejection, graft dysfunction, and survival at 1 year.
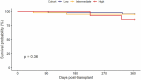
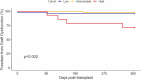
Results: Median age at time of transplant was 59.8 years, and 77.6% were men. In the dd-cfDNA group, there were significantly fewer episodes of EMBx defined acute cellular rejection (ACR) (2.5% vs 18.8%, p < 0.001) and treated ACR (4.2% vs 19.6%, p = 0.001). Comparatively, there were more EMBx defined antibody-mediated rejection (AMR) (5% vs 0.9%) and treated AMR (5% vs 2.7%) in the dd-cfDNA group. No significant differences were observed in graft dysfunction, presence of donor-specific antibodies, or survival at 1 year.
Conclusions: In conclusion, a modern rejection surveillance protocol utilizing noninvasive testing is safe, led to significantly fewer EMBx, fewer treated rejection episodes, and no difference in survival at 1 year. More AMR episodes identified via dd-cfDNA could lead the way for more accurate diagnostic and treatment decisions.
Keywords: Dd-cfDNA; GEP; HeartCare; heart transplant; rejection.
Conflict of interest statement
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Dr Shelley Hall is a consultant for CareDx and Natera. Other authors have no disclosures to report. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. Funding: None. Presented at The American Transplant Congress Annual Meeting, San Diego, California, June 3, 2023.
Figures



References
-
- Crespo-Leiro M.G., Zuckermann A., Bara C., et al. Concordance among pathologists in the second Cardiac Allograft Rejection Gene Expression Observational Study (CARGO II) Transplantation. 2012;94:1172–1177. - PubMed
-
- Deng M.C., Eisen H.J., Mehra M.R., et al. Noninvasive discrimination of rejection in cardiac allograft recipients using gene expression profiling. Am J Transplant. 2006;6:150–160. - PubMed
LinkOut - more resources
Full Text Sources

