Guselkumab binding to CD64+ IL-23-producing myeloid cells enhances potency for neutralizing IL-23 signaling
- PMID: 40145093
- PMCID: PMC11937023
- DOI: 10.3389/fimmu.2025.1532852
Guselkumab binding to CD64+ IL-23-producing myeloid cells enhances potency for neutralizing IL-23 signaling
Abstract
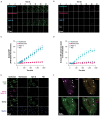
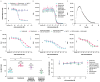
IL-23 is implicated in the pathogenesis of immune-mediated inflammatory diseases, and myeloid cells that express Fc gamma receptor 1 (FcγRI or CD64) on their surface have been recently identified as a primary source of IL-23 in inflamed tissue. Our complementary analyses of transcriptomic datasets from psoriasis and IBD showed increased expression of CD64 and IL-23 transcripts in inflamed tissue, and greater abundance of cell types with co-expression of CD64 and IL-23. These findings led us to explore potential implications of CD64 binding on the function of IL-23-targeting monoclonal antibodies (mAbs). Guselkumab and risankizumab are mAbs that target the IL-23p19 subunit. Guselkumab has a native Fc domain while risankizumab contains mutations that diminish binding to FcγRs. In flow cytometry assays, guselkumab, but not risankizumab, showed Fc-mediated binding to CD64 on IFNγ-primed monocytes. Guselkumab bound CD64 on IL-23-producing inflammatory monocytes and simultaneously captured IL-23 secreted from these cells. Guselkumab binding to CD64 did not induce cytokine production. In live-cell confocal imaging of CD64+ macrophages, guselkumab, but not risankizumab, mediated IL-23 internalization to low-pH intracellular compartments. Guselkumab and risankizumab demonstrated similar potency for inhibition of IL-23 signaling in cellular assays with exogenous addition of IL-23. However, in a co-culture of IL-23-producing CD64+ THP-1 cells with an IL-23-responsive reporter cell line, guselkumab demonstrated Fc-dependent enhanced potency compared to risankizumab for inhibiting IL-23 signaling. These in vitro data highlight the potential for guselkumab binding to CD64 in inflamed tissue to contribute to the potent neutralization of IL-23 at its cellular source.
Keywords: CD64; IL-23; IL-23p19 subunit inhibitors; RNA sequencing; guselkumab; immune-mediated inflammatory diseases; in vitro cellular assays; risankizumab.
Copyright © 2025 Sachen, Hammaker, Sarabia, Stoveken, Hartman, Leppard, Manieri, Bao, Greving, Lacy, DuPrie, Wertheimer, Deming, Brown, Hart, Li, Freeman, Keyes, Kohler, White, Karpowich, Steele, Elloso, Fakharzadeh, Goyal, Lavie, Abreu, Allez, Atreya, Bissonnette, Eyerich, Krueger, McGonagle, McInnes, Ritchlin and Fourie.
Conflict of interest statement
KS, DH, IS, BS, JH, KL, NM, PB, CG, EL, MD, JW, JD, JB, AH, HL, TF, BK, KK, IW, NK, RS, and SF are employees of and may hold stock in Johnson & Johnson. ME, KG, FL, and AF are former employees of Johnson & Johnson. MAb is a consultant or served on advisory boards for AbbVie, Arena Pharmaceuticals Inc., now Pfizer, Bristol Myers Squibb, Celsius Therapeutics, Gilead, Janssen Pharmaceuticals, Janssen Global Services, Lilly, Pfizer, Prometheus Biosciences, and UCB; and received fees for lecturing from Alimentiv, Janssen Pharmaceuticals, Prime, and WebMD Global LLC. MAl received consulting and/or lecture fees from AbbVie, Amgen, Biogen, Boehringer Ingelheim, Bristol Myers Squibb, Celgene, Celltrion Healthcare, Ferring, Genentech, Gilead, IQVIA, Janssen, Novartis, Pfizer, Roche, Takeda, and Tillotts; and received grant support from Genentech/Roche, Innate Pharma, Janssen, and Takeda. RA served as a paid or unpaid consultant for or received honoraria from AbbVie, Amgen, Arena Pharmaceuticals, Biogen, Boehringer Ingelheim, Bristol Myers Squibb, Celgene, Celltrion Healthcare, Dr. Falk Pharma, Ferring, Fresenius Kabi, Galapagos, Gilead, GSK, InDex Pharmaceuticals, Janssen, Kiniksa Pharmaceuticals, Merck Sharp & Dohme, Novartis, Pfizer, Roche, Samsung Bioepis, Stelic, sterna biologicals, Takeda, and Tillotts. RB is an advisory board member, consultant, speaker, and/or investigator for and received honoraria and/or grants from AbbVie, Alumis, Amgen, AnaptysBio, Bausch Health, Boston, Bristol Myer Squibb/Celgene, Dermavant, Janssen, LEO Pharma, Lilly, Nimbus Therapeutics, Novartis, Pfizer, Regeneron, UCB, Ventyx, and Xenco Medical; and is an employee and shareholder of Innovaderm Research. KE received speaker fees from, and/or served on advisory boards for AbbVie, Almirall, Boehringer Ingelheim, Bristol Myers Squibb, Hexal, Janssen, LEO Pharma, Lilly, Novartis, Pfizer, Sanofi, and UCB. JK served as a consultant or received honoraria for AbbVie, Aclaris Therapeutics, Allergan, Almirall, Amgen, Arena Pharmaceuticals, Aristea Therapeutics, Asana BioSciences, Aurigene, Biogen, Boehringer Ingelheim, Bristol Myers Squibb, Escalier Biosciences, Galapagos, Janssen, Lilly, MoonLake Immunotherapeutics, Nimbus Therapeutics, Novartis, Pfizer, Sanofi, Sienna Biopharmaceuticals, Sun Pharma, Target-Derm, UCB, Valeant, and Ventyx. DM received research grants from AbbVie, BMS, Celgene, Janssen, Lilly, Merck, MoonLake Immunotherapeutics, Novartis, Pfizer, and UCB; received honoraria or consultation fees from AbbVie, Celgene, Janssen, Merck, Novartis, Pfizer, and UCB; and participated in speakers bureau for AbbVie, Celgene, Janssen, Merck, Novartis, Pfizer, and UCB. IM received consultant fees from AbbVie, Amgen, AstraZeneca, Bristol Myers Squibb, Cabaletta Bio, Compugen, Gilead, GSK, Janssen, Lilly, Novartis, Pfizer, Roche, Sanofi, and UCB; received grant/research support from Amgen, AstraZeneca, Bristol Myers Squibb, GSK, Janssen, Lilly, Novartis, Roche, and UCB; is a shareholder for Causeway, Compugen, and Evelo; is a board member for the National Health Service Greater Glasgow and Clyde; is on the board of directors for Evelo; and is a trustee for Versus Arthritis. CR received grant/research support from AbbVie, Amgen, and UCB; and received consulting fees from AbbVie, Amgen, Gilead, Janssen, Lilly, Novartis, Pfizer, and UCB.
Figures



Comment in
-
Commentary: Guselkumab binding to CD64+ IL-23-producing myeloid cells enhances potency for neutralizing IL-23 signaling.Front Immunol. 2025 May 20;16:1604337. doi: 10.3389/fimmu.2025.1604337. eCollection 2025. Front Immunol. 2025. PMID: 40463376 Free PMC article. No abstract available.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

