High Sugar Induced RCC2 Lactylation Drives Breast Cancer Tumorigenicity Through Upregulating MAD2L1
- PMID: 40145796
- PMCID: PMC12140329
- DOI: 10.1002/advs.202415530
High Sugar Induced RCC2 Lactylation Drives Breast Cancer Tumorigenicity Through Upregulating MAD2L1
Abstract
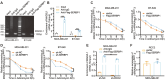
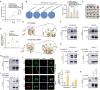
Lactylation is a novel post-translational modification mediated by lactate, widely present in the lysine residues of both histone and non-histone proteins. However, the specific regulatory mechanisms and downstream target proteins remain unclear. Herein, it is demonstrated that the RCC2 protein may serve as a critical link between material metabolism and cell division, promoting the rapid proliferation of breast cancer under high glucose conditions. Mechanistically, the activation of glycolysis leads to an increase in lactate. Then, acyltransferase KAT2A mediates RCC2 lactylation at K124, which assists RCC2 in recruiting free SERBP1, thereby stabilizing MAD2L1 mRNA. The lactylation of RCC2 mediates the activation of the cellular MAD2L1 signaling pathway and contributes to the progression of breast cancer. A small molecule inhibitor slows down cell proliferation by binding to the RCC2 active pocket and specifically blocking RCC2 lactylation. The findings elucidate the mechanism behind the upregulation of MAD2L1 in murine tumors associated with a high-sugar diet as reported in prior study and suggest a novel therapeutic strategy of targeting RCC2 lactylation to restrict the rapid proliferation of breast cancer cell in a high-lactate microenvironment.
Keywords: MAD2L1; RCC2; SERBP1; cell division; high sugar diet.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Giaquinto A. N., Sung H., Miller K. D., Kramer J. L., Newman L. A., Minihan A., Jemal A., Siegel R. L., CA Cancer J. Clin. 2022. - PubMed
-
- Aftimos P., Oliveira M., Irrthum A., Fumagalli D., Sotiriou C., Gal‐Yam E. N., Robson M. E., Ndozeng J., Di Leo A., Ciruelos E. M., de Azambuja E., Viale G., Scheepers E. D., Curigliano G., Bliss J. M., Reis‐Filho J. S., Colleoni M., Balic M., Cardoso F., Albanell J., Duhem C., Marreaud S., Romagnoli D., Rojas B., Gombos A., Wildiers H., Guerrero‐Zotano A., Hall P., Bonetti A., Larsson K. F., et al., Cancer Discovery 2021, 11, 2796. - PMC - PubMed
-
- Koppenol W. H., Bounds P. L., Dang C. V., Nat. Rev. Cancer 2011, 11, 325. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
