A patient-derived T cell lymphoma biorepository uncovers pathogenetic mechanisms and host-related therapeutic vulnerabilities
- PMID: 40147445
- PMCID: PMC12047492
- DOI: 10.1016/j.xcrm.2025.102029
A patient-derived T cell lymphoma biorepository uncovers pathogenetic mechanisms and host-related therapeutic vulnerabilities
Abstract
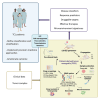
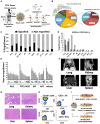
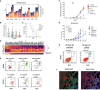
Peripheral T cell lymphomas (PTCLs) comprise heterogeneous malignancies with limited therapeutic options. To uncover targetable vulnerabilities, we generate a collection of PTCL patient-derived tumor xenografts (PDXs) retaining histomorphology and molecular donor-tumor features over serial xenografting. PDX demonstrates remarkable heterogeneity, complex intratumor architecture, and stepwise trajectories mimicking primary evolutions. Combining functional transcriptional stratification and multiparametric imaging, we identify four distinct PTCL microenvironment subtypes with prognostic value. Mechanistically, we discover a subset of PTCLs expressing Epstein-Barr virus-specific T cell receptors and uncover the capacity of cancer-associated fibroblasts of counteracting treatments. PDXs' pre-clinical testing captures individual vulnerabilities, mirrors donor patients' clinical responses, and defines effective patient-tailored treatments. Ultimately, we assess the efficacy of CD5KO- and CD30- Chimeric Antigen Receptor T Cells (CD5KO-CART and CD30_CART, respectively), demonstrating their therapeutic potential and the synergistic role of immune checkpoint inhibitors for PTCL treatment. This repository represents a resource for discovering and validating intrinsic and extrinsic factors and improving the selection of drugs/combinations and immune-based therapies.
Keywords: CAR-T; T cell lymphoma; clonal evolution; drug screenings; microenvironment; patient-derived tumor xenografts; pre-clinical trials; precision medicine; repository; stratification.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests D.M.W. is an employee of Merck and has an equity interest in Ajax, Bantam, and Travera. F.B. receives institutional research funds from ADC Therapeutics, Bayer AG, Cellestia, Helsinn, HTG Molecular Diagnostics, ImmunoGen, iOnctura, Menarini Ricerche, NEOMED Therapeutics 1, Nordic Nanovector ASA, and Spexis AG; advisory board fees from Novartis; consultancy fee from Helsinn and Menarini; and travel grants from Amgen, AstraZeneca, and iOnctura and provided expert statements to HTG Molecular Diagnostics.
Figures






References
-
- Mak V., Hamm J., Chhanabhai M., Shenkier T., Klasa R., Sehn L.H., Villa D., Gascoyne R.D., Connors J.M., Savage K.J. Survival of patients with peripheral T-cell lymphoma after first relapse or progression: spectrum of disease and rare long-term survivors. J. Clin. Oncol. 2013;31:1970–1976. doi: 10.1200/JCO.2012.44.7524. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

