Medium from human iPSC-derived primitive macrophages promotes adult cardiomyocyte proliferation and cardiac regeneration
- PMID: 40148355
- PMCID: PMC11950653
- DOI: 10.1038/s41467-025-58301-8
Medium from human iPSC-derived primitive macrophages promotes adult cardiomyocyte proliferation and cardiac regeneration
Abstract
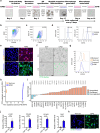
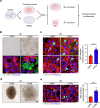
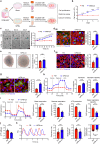
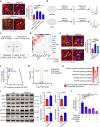
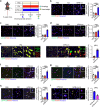
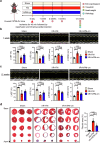
Heart injury has been characterized by the irreversible loss of cardiomyocytes comprising the contractile tissues of the heart and thus strategies enabling adult cardiomyocyte proliferation are highly desired for treating various heart diseases. Here, we test the ability of human induced pluripotent stem cell-derived primitive macrophages (hiPMs) and their conditioned medium (hiPM-cm) to promote human cardiomyocyte proliferation and enhance cardiac regeneration in adult mice. We find that hiPMs promote human cardiomyocyte proliferation, which is recapitulated by hiPM-cm through the activation of multiple pro-proliferative pathways, and a secreted proteome analysis identifies five proteins participating in this activation. Subsequent in vivo experiments show that hiPM-cm promotes adult cardiomyocyte proliferation in mice. Lastly, hiPM-cm enhances cardiac regeneration and improves contractile function in injured adult mouse hearts. Together, our study demonstrates the efficacy of using hiPM-cm in promoting adult cardiomyocyte proliferation and cardiac regeneration to serve as an innovative treatment for heart disease.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures

References
-
- Braunwald, E. The war against heart failure: the Lancet lecture. Lancet385, 812–824 (2015). - PubMed
-
- Chen, Y. et al. Reversible reprogramming of cardiomyocytes to a fetal state drives heart regeneration in mice. Science373, 1537–1540 (2021). - PubMed
-
- Humphries, S., Mars, K., Hofmann, R., Held, C. & Olsson, E. M. G. Randomized evaluation of routine beta-blocker therapy after myocardial infarction quality of life (RQoL): design and rationale of a multicentre, prospective, randomized, open, blinded endpoint study. Eur. Heart J. Open3, oead036 (2023). - PMC - PubMed
-
- Steg, P. G. Routine beta-blockers in secondary prevention - on injured reserve. N. Engl. J. Med.390, 1434–1436 (2024). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

