The transcription factor LHX2 mediates and enhances oncogenic BMP signaling in medulloblastoma
- PMID: 40148468
- PMCID: PMC12501261
- DOI: 10.1038/s41418-025-01488-6
The transcription factor LHX2 mediates and enhances oncogenic BMP signaling in medulloblastoma
Abstract
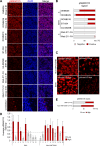
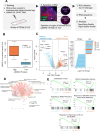
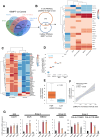
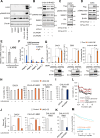
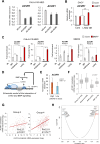
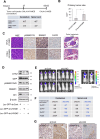
Oncogenic events perturb cerebellar development leading to medulloblastoma, a common childhood brain malignancy. Molecular analyses classify medulloblastoma into the WNT, SHH, Group 3 and Group 4 subgroups. Bone morphogenetic protein (BMP) pathways control cerebellar development and have been linked to the progression of medulloblastoma disease, with major remaining gaps in their mechanistic and clinically-relevant roles. We therefore aimed at exploring BMP mechanisms of action in medulloblastoma. Patient-derived tumors from different subgroups were analyzed in mouse xenografts, complemented by independent tumor immunohistochemical analysis. Cell-based assays analyzed signaling mechanisms. Medulloblastoma cell orthotopic xenografts analyzed tumor growth and metastasis in vivo. Active BMP signaling, detected as nuclear and phosphorylated SMAD1/5, characterized several medulloblastoma subgroups, with enrichment in Group 4, SHH and Group 3 tumors. Spatial transcriptomics in tumor areas, complemented by transcriptomic analysis of multiple cell models, identified BMP-dependent transcriptional induction of the LIM-homeobox gene 2 (LHX2). BMP signaling via SMADs induced LHX2 expression and LHX2 transcriptionally induced BMP type I receptor (ACVR1) expression by association with the proximal promoter region of the ACVR1 gene. BMP signaling and LHX2 gain-of-function expression led to enriched stemness and associated chemoresistance in medulloblastoma cultures. In-mouse orthotopic transplantation of paired primary/recurrent Group 4 medulloblastoma cell populations, correspondingly expressing LHX2-low/BMP-low signaling and LHX2-high/BMP-high signaling, ascribed to the latter (high) group more efficient tumor propagation and spinal cord metastatic potential. Depletion of LHX2 in these recurrent tumor cells suppressed both BMP signaling and tumor propagation in vivo. Thus, LHX2 cooperates with, and enhances, oncogenic BMP signaling in medulloblastoma tumors. The molecular pathway that couples LHX2 function to BMP signaling in medulloblastoma deepens our understanding this malignancy.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval and consent to participate: All the procedures were performed in accordance with the guidelines for Ethical Conduct in the Care and Use of Animals as stated in The International Guiding Principles for Biomedical Research involving Animals, developed by the Council for International Organizations of Medical Sciences (CIOMS). For the animal experiments in Sweden, mouse husbandry and manipulations followed the national guidelines after approval by the Uppsala University Animal Experiment Ethics Board (C105/16, 5.8.18-16350/2017 and 5.8.18-18303/2021). For the animal experiments in Spain, mice were housed according to institutional guidelines and all experimental procedures were performed in compliance with the guidelines for the welfare of experimental animals as approved by the Universidad of Zaragoza Ethics Committee (CEICA PI34/21).
Figures
References
MeSH terms
Substances
Grants and funding
- PR2018-0091, PR2020-0088, PR2023-0115/Barncancerfonden (Swedish Childhood Cancer Foundation)
- TJ2021-0078/Barncancerfonden (Swedish Childhood Cancer Foundation)
- Uppsala Branch/Ludwig Institute for Cancer Research (Ludwig Cancer Research)
- Overseas Research Fellowship/MEXT | Japan Society for the Promotion of Science (JSPS)
- GA21-JPN-0018/Sasakawa Memorial Health Foundation (SMHF)
- 22 0555 01H/Cancerfonden (Swedish Cancer Society)
- FO2020-0335/Stiftelsen Lars Hiertas Minne (Lars Hierta Memorial Foundation)
- 2021-04284, 2022-211/Magnus Bergvalls Stiftelse (Magnus Bergvall Foundation)
- 787472/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- 2020-01291/Vetenskapsrådet (Swedish Research Council)
LinkOut - more resources
Full Text Sources
Research Materials

