DYRK1A inhibition results in MYC and ERK activation rendering KMT2A-R acute lymphoblastic leukemia cells sensitive to BCL2 inhibition
- PMID: 40148558
- PMCID: PMC12055583
- DOI: 10.1038/s41375-025-02575-w
DYRK1A inhibition results in MYC and ERK activation rendering KMT2A-R acute lymphoblastic leukemia cells sensitive to BCL2 inhibition
Abstract
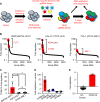
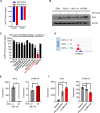
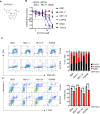
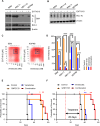
Unbiased kinome-wide CRISPR screening identified DYRK1A as a potential therapeutic target in KMT2A-rearranged (KMT2A-R) B-acute lymphoblastic leukemia (ALL). Mechanistically, we demonstrate that DYRK1A is regulated by the KMT2A fusion protein and affects cell proliferation by regulating MYC expression and ERK phosphorylation. We further observed that pharmacologic DYRK1A inhibition markedly reduced human KMT2A-R ALL cell proliferation in vitro and potently decreased leukemia proliferation in vivo in drug-treated patient-derived xenograft mouse models. DYRK1A inhibition induced expression of the proapoptotic factor BIM and reduced the expression of BCL-XL, consequently sensitizing KMT2A-R ALL cells to BCL2 inhibition. Dual inhibition of DYRK1A and BCL2 synergistically decreased KMT2A-R ALL cell survival in vitro and reduced leukemic burden in mice. Taken together, our data establishes DYRK1A as a novel therapeutic target in KMT2A-R ALL and credential dual inhibition of DYRK1A and BCL2 as an effective translational therapeutic strategy for this high-risk ALL subtype.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: JDC is on the SAB for Alethiomics and receives research funding from Syndax. TAM is a paid consultant for and shareholder in Dark Blue Therapeutics Ltd. SKT receives research funding from Incyte Corporation and Kura Oncology, serves/d on scientific advisory boards for Aleta Biotherapeutics, AstraZeneca, Jazz Pharmaceuticals, Kura Oncology, Syndax Pharmaceuticals, and Wugen, Inc., and has received travel support from Amgen and Jazz Pharmaceuticals (all for unrelated studies). Ethics approval and consent to participate: All methods were performed in accordance with the relevant guidelines and regulations. We have complied with all the relevant ethical regulations for animal testing and research. All animal-related experiments have received ethical approval from Loma Linda University Institutional Animal Care and Use Committee (IACUC-23-029).
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

