Inducible transposon mutagenesis identifies bacterial fitness determinants during infection in mice
- PMID: 40148565
- PMCID: PMC12055562
- DOI: 10.1038/s41564-025-01975-z
Inducible transposon mutagenesis identifies bacterial fitness determinants during infection in mice
Abstract
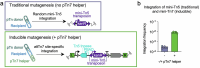
Transposon insertion sequencing (Tn-seq) is a powerful method for genome-scale forward genetics in bacteria. However, inefficient transposon delivery or stochastic loss of mutants due to population bottlenecks can limit its effectiveness. Here we have developed 'InducTn-seq', where an arabinose-inducible Tn5 transposase enables temporal control of mini-Tn5 transposition. InducTn-seq generated up to 1.2 million transposon mutants from a single colony of enterotoxigenic Escherichia coli, Salmonella typhimurium, Shigella flexneri and Citrobacter rodentium. This mutant diversity enabled more sensitive detection of subtle fitness defects and measurement of quantitative fitness effects for essential and non-essential genes. Applying InducTn-seq to C. rodentium in a mouse model of infectious colitis bypassed a highly restrictive host bottleneck, generating a diverse population of >5 × 105 unique transposon mutants compared to 10-102 recovered by traditional Tn-seq. This in vivo screen revealed that the C. rodentium type I-E CRISPR system is required to suppress a toxin otherwise activated during gut colonization. Our findings highlight the potential of InducTn-seq for genome-scale forward genetic screens in bacteria.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures







Update of
-
Inducible transposon mutagenesis for genome-scale forward genetics.bioRxiv [Preprint]. 2024 May 21:2024.05.21.595064. doi: 10.1101/2024.05.21.595064. bioRxiv. 2024. Update in: Nat Microbiol. 2025 May;10(5):1171-1183. doi: 10.1038/s41564-025-01975-z. PMID: 38826325 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- P30DK034854/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- T32DK007477-37/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- F31AI156949/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01AI042347/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
LinkOut - more resources
Full Text Sources
Research Materials

