The Trade-Off Between Sanitizer Resistance and Virulence Genes: Genomic Insights into E. coli Adaptation
- PMID: 40149102
- PMCID: PMC11939141
- DOI: 10.3390/antibiotics14030291
The Trade-Off Between Sanitizer Resistance and Virulence Genes: Genomic Insights into E. coli Adaptation
Abstract
Background: Escherichia coli is one of the most studied bacteria worldwide due to its genetic plasticity. Recently, in addition to characterizing its pathogenic potential, research has focused on understanding its resistance profile to inhibitory agents, whether these be antibiotics or sanitizers.
Objectives: The present study aimed to investigate six of the main serogroups of foodborne infection (O26, O45, O103, O111, O121, and O157) and to understand the dynamics of heterogeneity in resistance to sanitizers derived from quaternary ammonium compounds (QACs) and peracetic acid (PAA) using whole-genome sequencing (WGS).
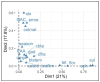
Methods: Twenty-four E. coli strains with varied resistance profiles to QACs and PAA were analyzed by WGS using NovaSeq6000 (150 bp Paired End reads). Bioinformatic analyses included genome assembly (Shovill), annotation via Prokka, antimicrobial resistance gene identification using Abricate, and core-genome analysis using Roary. A multifactorial multiple correspondence analysis (MCA) was conducted to explore gene-sanitizer relationships. In addition, a large-scale analysis utilizing the NCBI Pathogen Detection database involved a 2 × 2 chi-square test to examine associations between the presence of qac and stx genes.
Results: The isolates exhibited varying antimicrobial resistance profiles, with O45 and O157 being the most resistant serogroups. In addition, the qac gene was identified in only one strain (S22), while four other strains carried the stx gene. Through multifactorial multiple correspondence analysis, the results obtained indicated that strains harboring genes encoding Shiga toxin (stx) presented profiles that were more likely to be sensitive to QACs. To further confirm these results, we analyzed 393,216 E. coli genomes from the NCBI Pathogen Detection database. Our results revealed a significant association (p < 0.001) between the presence of qac genes and the absence of stx1, stx2, or both toxin genes.
Conclusion: Our findings highlight the complexity of bacterial resistance mechanisms and suggest that non-pathogenic strains may exhibit greater tolerance to QAC sanitizer than those carrying pathogenicity genes, particularly Shiga toxin genes.
Keywords: bacterial communities; bacteriophage; evolutionary mechanisms; foodborne bacteria; sanitizer resistance.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Similar articles
-
Isolation and characterization of shiga toxin-producing escherichia coli serogroups O26, O45, O103, O111, O113, O121, O145, and O157 shed from range and feedlot cattle from postweaning to slaughter.J Food Prot. 2014 Jul;77(7):1052-61. doi: 10.4315/0362-028X.JFP-13-373. J Food Prot. 2014. PMID: 24988009
-
Summer and Winter Prevalence of Shiga Toxin-Producing Escherichia coli (STEC) O26, O45, O103, O111, O121, O145, and O157 in Feces of Feedlot Cattle.Foodborne Pathog Dis. 2015 Aug;12(8):726-32. doi: 10.1089/fpd.2015.1987. Epub 2015 Jun 15. Foodborne Pathog Dis. 2015. PMID: 26075548
-
DNA Microarray-Based Genomic Characterization of the Pathotypes of Escherichia coli O26, O45, O103, O111, and O145 Isolated from Feces of Feedlot Cattle †.J Food Prot. 2019 Mar;82(3):395-404. doi: 10.4315/0362-028X.JFP-18-393. J Food Prot. 2019. PMID: 30794460
-
Diversity of CRISPR loci and virulence genes in pathogenic Escherichia coli isolates from various sources.Int J Food Microbiol. 2015 Jul 2;204:41-6. doi: 10.1016/j.ijfoodmicro.2015.03.025. Epub 2015 Mar 27. Int J Food Microbiol. 2015. PMID: 25839984
-
Shiga Toxin-Producing Escherichia coli in Feces of Finisher Pigs: Isolation, Identification, and Public Health Implications of Major and Minor Serogroups†.J Food Prot. 2021 Jan 1;84(1):169-180. doi: 10.4315/JFP-20-329. J Food Prot. 2021. PMID: 33411931
References
-
- de Assis D.C.S., da Silva T.M.L., Brito R.F., da Silva L.C.G., Lima W.G., Brito J.C.M. Shiga toxin-producing Escherichia coli (STEC) in bovine meat and meat products over the last 15 years in Brazil: A systematic review and meta-analysis. Meat Sci. 2021;173:108394. doi: 10.1016/j.meatsci.2020.108394. - DOI - PubMed
-
- Ludwig J.B., Shi X., Shridhar P.B., Roberts E.L., DebRoy C., Phebus R.K., Bai J., Nagaraja T.G. Multiplex PCR assays for the detection of one hundred and thirty-seven serogroups of shiga toxin-producing Escherichia coli associated with cattle. Front. Cell. Infect. Microbiol. 2020;10:378. doi: 10.3389/fcimb.2020.00378. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

