Oncological Treatment Adverse Reaction Prediction: Development and Initial Validation of a Pharmacogenetic Model in Non-Small-Cell Lung Cancer Patients
- PMID: 40149417
- PMCID: PMC11942520
- DOI: 10.3390/genes16030265
Oncological Treatment Adverse Reaction Prediction: Development and Initial Validation of a Pharmacogenetic Model in Non-Small-Cell Lung Cancer Patients
Abstract
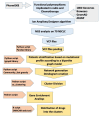
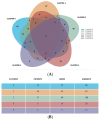
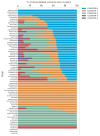
Background/Objectives: The accurate prediction of adverse drug reactions (ADRs) to oncological treatments still poses a clinical challenge. Chemotherapy is usually selected based on clinical trials that do not consider patient variability in ADR risk. Consequently, many patients undergo multiple treatments to find the appropriate medication or dosage, enhancing ADR risks and increasing the chance of discontinuing therapy. We first aimed to develop a pharmacogenetic model for predicting chemotherapy-induced ADRs in cancer patients (the ANTIBLASTIC DRUG MULTIPANEL PLATFORM) and then to assess its feasibility and validate this model in patients with non-small-cell lung cancer (NSCLC) undergoing oncological treatments. Methods: Seventy NSCLC patients of all stages that needed oncological treatment at our facility were enrolled, reflecting the typical population served by our institution, based on geographic and demographic characteristics. Treatments followed existing guidelines, and patients were continuously monitored for adverse reactions. We developed and used a multipanel platform based on 326 SNPs that we identified as strongly associated with response to cancer treatments. Subsequently, a network-based algorithm to link these SNPs to molecular and biological functions, as well as efficacy and adverse reactions to oncological treatments, was used. Results: Data and blood samples were collected from 70 NSCLC patients. A bioinformatic analysis of all identified SNPs highlighted five clusters of patients based on variant aggregations and the associated genes, suggesting potential susceptibility to treatment-related toxicity. We assessed the feasibility of the platform and technically validated it by comparing NSCLC patients undergoing the same course of treatment with or without ADRs against the cluster combination. An odds ratio analysis confirmed the correlation between cluster allocation and increased ADR risk, indicating specific treatment susceptibilities. Conclusions: The ANTIBLASTIC DRUG MULTIPANEL PLATFORM was easily applicable and able to predict ADRs in NSCLC patients undergoing oncological treatments. The application of this novel predictive model could significantly reduce adverse drug reactions and improve the rate of chemotherapy completion, enhancing patient outcomes and quality of life. Its potential for broader prescription management suggests significant treatment improvements in cancer patients.
Keywords: cancer; individual variability; oncology; personalized medicine; pharmacogenetics; pharmacogenomics; supportive care; symptom management.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Caudle K.E., Klein T.E., Hoffman J.M., Muller D.J., Whirl-Carrillo M., Gong L., McDonagh E.M., Sangkuhl K., Thorn C.F., Schwab M., et al. Incorporation of pharmacogenomics into routine clinical practice: The Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline development process. Curr. Drug Metab. 2014;15:209–217. doi: 10.2174/1389200215666140130124910. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

