The Transformation Experiment of Frederick Griffith I: Its Narrowing and Potential for the Creation of Novel Microorganisms
- PMID: 40150788
- PMCID: PMC11939280
- DOI: 10.3390/bioengineering12030324
The Transformation Experiment of Frederick Griffith I: Its Narrowing and Potential for the Creation of Novel Microorganisms
Abstract
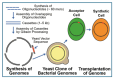
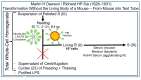
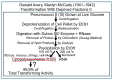
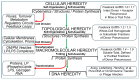
The construction of artificial microorganisms often relies on the transfer of genomes from donor to acceptor cells. This synthetic biology approach has been considerably fostered by the J. Craig Venter Institute but apparently depends on the use of microorganisms, which are very closely related. One reason for this limitation of the "creative potential" of "classical" transformation is the requirement for adequate "fitting" of newly synthesized polypeptide components, directed by the donor genome, to interacting counterparts encoded by the pre-existing acceptor genome. Transformation was introduced in 1928 by Frederick Griffith in the course of the demonstration of the instability of pneumococci and their conversion from rough, non-pathogenic into smooth, virulent variants. Subsequently, this method turned out to be critical for the identification of DNA as the sole matter of inheritance. Importantly, the initial experimental design (1.0) also considered the inheritance of both structural (e.g., plasma membranes) and cybernetic information (e.g., metabolite fluxes), which, in cooperation, determine topological and cellular heredity, as well as fusion and blending of bacterial cells. In contrast, subsequent experimental designs (1.X) were focused on the use of whole-cell homogenates and, thereafter, of soluble and water-clear fractions deprived of all information and macromolecules other than those directing protein synthesis, including outer-membrane vesicles, bacterial prions, lipopolysaccharides, lipoproteins, cytoskeletal elements, and complexes thereof. Identification of the reasons for this narrowing may be helpful in understanding the potential of transformation for the creation of novel microorganisms.
Keywords: cellular heredity; horizontal gene transfer; science and technology studies; scientific reductionism; synthetic biology; transforming principle.
Conflict of interest statement
The author declares no conflict of interest.
Figures













References
-
- Gibson D.G., Benders G.A., Andrews-Pfannkoch C., Denisova E.A., Baden-Tillson H., Zaveri J., Stockwell T.B., Brownley A., Thomas D.W., Algire M.A., et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. - DOI - PubMed
-
- Lartigue C., Vashee S., Algire M.A., Chuang R.-Y., Benders G.A., Ma L., Noskov V.N., Denisova E.A., Gibson D.G., Assad-Garcia N., et al. Creating bacterial strains from genomes that have been cloned and engineered in yeast. Science. 2009;325:1693–1696. - PubMed
-
- Gibson D.G., Glass J.I., Lartigue C., Noskov V.N., Chuang R.-Y., Algire M.A., Benders G.A., Montague M.G., Ma L., Moodie M.M., et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. - PubMed
-
- Hutchison C.A., 3rd, Chuang R.-A., Noskov V.N., Assad-Garcia N., Deerinck T.J., Ellisman M.H., Gill J., Kannan K., Karas B.J., Ma L., et al. Design and synthesis of a minimal genome. Science. 2016;351:aad6253. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

