UGDH Lactylation Aggravates Osteoarthritis by Suppressing Glycosaminoglycan Synthesis and Orchestrating Nucleocytoplasmic Transport to Activate MAPK Signaling
- PMID: 40150862
- PMCID: PMC12120796
- DOI: 10.1002/advs.202413709
UGDH Lactylation Aggravates Osteoarthritis by Suppressing Glycosaminoglycan Synthesis and Orchestrating Nucleocytoplasmic Transport to Activate MAPK Signaling
Abstract
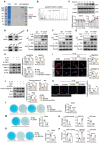
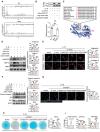
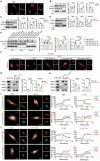
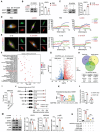
Osteoarthritis (OA) progression is closely related to dysregulated glycolysis. As the primary metabolite of glycolysis, lactate plays a detrimental role in OA. However, how lactate exacerbates OA process remains unclear. Here, this study revealed that lactate levels are elevated in the synovial fluid of OA patients and IL-1β-treated human primary chondrocytes, promoting protein pan-lactylation. Functionally, hyper-lactylation exacerbates chondrocytes extracellular matrix (ECM) degradation and cell apoptosis in vitro and in vivo. Moreover, UDP-glucose dehydrogenase (UGDH) is proven to be the key lactylated protein in lactate-treated chondrocytes, which undergoes lactylation at lysine 6 (K6). Lactylated UGDH repressed its enzymatic activity, reducing glycosaminoglycan synthesis and disregulating its nuclear-cytoplasmic distribution. Mechanistically, K6 lactylation of UGDH impedes the interaction of UGDH and signal transducer and activator of transcription 1 (STAT1), thus promoting the transcription of mitogen-activated protein kinase kinase kinase 8 (MAP3K8) and activating the MAPK signaling pathway. Importantly, in vitro and in vivo treatment with A485, a specific acyltransferase P300 inhibitor, suppressed UGDH lactylation and rescued chondrocytes ECM degradation and OA progression. These findings uncover a new mechanism underlying OA pathogenesis and highlight the potential of targeting UGDH lactylation as a novel therapeutic strategy for OA.
Keywords: chondrocytes; lactate; lactylation; osteoarthritis; udp‐glucose dehydrogenase.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Duong V., Oo W. M., Ding C., Culvenor A. G., Hunter D. J., JAMA, J. Am. Med. Assoc. 2023, 330, 1568. - PubMed
-
- Hunter D. J., Osteoarthritis B.‐Z. S., Lancet 2019, 393, 1745. - PubMed
-
- Herrero‐Beaumont G., Pérez‐Baos S., Sánchez‐Pernaute O., Roman‐Blas J. A., Lamuedra A., Largo R., Biochem. Pharmacol. 2019, 165, 24. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
