Discovery of De Novo Macrocycle Inhibitors of Histone Deacetylase 11
- PMID: 40151233
- PMCID: PMC11938020
- DOI: 10.1021/jacsau.4c01148
Discovery of De Novo Macrocycle Inhibitors of Histone Deacetylase 11
Abstract
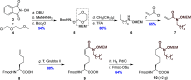
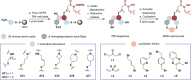
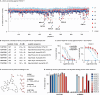
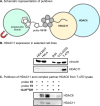
Histone deacetylase (HDAC) enzymes are epigenetic regulators that affect diverse protein function by removing acyl groups from lysine side chains throughout the proteome. The most recently discovered human isozyme, HDAC11, differs from other HDACs in substrate preference and tissue expression profile. Elucidation of the biological function of this enzyme has been scarce and only a few chemical probes to help advance this insight have been developed thus far. Here we discovered macrocyclic inhibitors that exhibit selectivity for HDAC11 and penetrate the cytoplasmic membrane in cultured cells as determined by the chloroalkane penetration assay. Our work establishes the combination of de novo macrocycle synthesis with incorporation of N-alkylated hydroxamic acid moieties as a viable strategy for targeting HDAC11. Further, this study demonstrates the potential of applying macrocyclic peptide-based library synthesis to directly furnish high-affinity, cell-permeating ligands. The discovered inhibitors comprise tool compounds for the investigation of the biological function of HDAC11.
© 2025 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare the following competing financial interest(s): EPFL has filed a PCT application covering parts of the presented library preparation method (WO/2022/242993) with A.L.N. and C.H. as co-inventors. C.H. is a co-founder of Orbis Medicines. The remaining authors declare no competing interests.
Figures



References
LinkOut - more resources
Full Text Sources
